Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Sheela Ramamoorthy.
Porcine circovirus type 2 (PCV2), the causative agent of a wasting disease in weanling piglets, has periodically evolved into several new subtypes since its discovery, indicating that the efficacy of current vaccines can be improved. Although a DNA virus, the mutation rates of PCV2 resemble RNA viruses. By mutating selected serine and leucine codons to increase the probability that they would change to stop codons during replication, rapid attenuation of the vaccine construct was achieved. In a weanling piglet model, the suicidal vaccine was highly effective and safe, providing proof of concept for a novel rapid attenuation approach that is broadly applicable to newly emerging RNA and DNA viruses.
- porcine
- circoviruses
- PCV2
- immunity
- cell-mediated immunity
- innate immunity
- antibody
- vaccine
- evolution
1. Introduction
In the last few decades, the number of newly emerging or re-emerging viral infections has increased significantly. Over 80% of these emerging or re-emerging infections are caused by RNA viruses. The highly mutable nature of pathogenic RNA viruses and the associated antigenic and genetic diversity is a long-standing challenge for effective vaccine development [1,2][1][2]. While much attention has been focused on approaches for developing rapid attenuation methods for RNA viruses, ssDNA viruses, such as porcine circovirus type 2 (PCV2), also have a high frequency of mutation. In general, attenuated vaccines are more effective and provide a longer duration of immunity than inactivated vaccines. However, achieving the optimal balance between safety and efficacy is a challenge in developing attenuated vaccines [1]. The long lead development time associated with conventional methods of viral attenuation and the knowledge of well-characterized virulence determinants being a pre-requisite for rational reverse genetics-based approaches hinder rapid-response vaccine development for emerging viruses [1,2][1][2]. Therefore, the availability of broadly applicable vaccine platforms that are safe and effective is a critical need. We have previously demonstrated that a minimally replicative vaccine against an enteric swine coronavirus [3] is highly effective at priming the immune system but is also quickly eliminated from the vaccinated host. Thus, minimally replicative vaccines can provide a much-desired amalgamation of vaccine safety and efficacy properties.
Porcine circovirus type 2 (PCV2) is a small, circular, single-stranded DNA virus that emerged in the mid-1990s as the cause of post-weaning multisystemic wasting disease syndrome (PMWS) in weanling pigs. Thereafter, several other clinical manifestations involving the respiratory, enteric, reproductive, and immune systems were attributed to PCV2 and collectively named porcine circovirus-associated diseases (PCVAD) [4]. Currently available commercial vaccines against PCV2 consist of inactivated and subunit vaccines that target the first detected PCV2a subtype and were introduced in the U.S. in 2006. They are highly effective in controlling clinical PCVAD and reducing the economic impacts of PCVAD but do not prevent infection or viral transmission [5,6][5][6]. The selection and emergence of variants that adapt to pre-existing immunity induced by either prior infection or vaccination is a well-documented phenomenon of several viruses, including PCV2 [6[6][7][8],7,8], canine parvovirus [9], hepatitis B virus [10], Japanese encephalitis virus [11], and Marek’s disease virus [12]. Similarly, since the discovery of PCV2, periodical shifts in the circulating subtypes have occurred, with the first detected PCV2a subtype being replaced by PCV2b, and more recently by the PCV2d subtype, which is now the predominating field strain [13], suggesting that the exploration of alternative approaches to vaccination is warranted [8]. Current PCV2 vaccines contain either the recombinant subunit PCV2a capsid protein or the whole inactivated virus and are extremely safe. However, live attenuated vaccine approaches can potentially improve the breadth and threshold of protection elicited by currently available vaccines and help to eradicate PCV2 in the long term [5] but are traditionally not favored due to safety concerns such as reversion to virulence or recombination with field strains. In addition, PCV2 vaccines with built-in capabilities for monitoring vaccine compliance in the field are unavailable but have an added advantage for producers as vaccine coverage can be monitored, and herd immunity targeted effectively. Thus, the objective of this study was to develop a rapid attenuation strategy targeting suicidal viral replication for PCV2, with the goal of achieving high vaccine safety and efficacy margins; and to enable serological monitoring of PCV2 vaccine compliance by the expression of a foreign epitope tag.
While the genetic instability of RNA viruses is a well-recognized problem for effective prevention, in general, DNA viruses are considered to be more genetically stable. However, single-stranded (ssDNA) DNA viruses, especially the circular rep-encoding single-stranded DNA viruses (CRESS viruses), including PCV2 [14], have rates of mutation that range from 10−3 to 10−6 substitutions/site/year, which are comparable to RNA viruses. With the availability of metagenomic sequencing technology, two newly emerging PCV types, PCV3 and PCV4, were recently detected in clinical cases of reproductive failure, systemic inflammation, myocarditis, and porcine dermatitis and nephropathy syndrome (PDNS) [15]. Thus, ssDNA viruses are genetically diverse, can have a broad host range [16[16][17],17], and potentially adapt to new hosts following mutational events [18].
Previously established methods for viral attenuation involving recoding the viral genes include codon deoptimization, codon pair deoptimization, and introduction of a CpG/UpA dinucleotide bias. These methods destabilize the protein translation process to reduce viral fitness while retaining antigenicity through the preservation of amino acid sequence [19]. However, they involve a significant alteration to the viral genetic composition and dinucleotide composition and can potentially alter the innate immune responses that are triggered by the sensing of viral nucleic acids [20]. As serine and leucine have the highest codon redundancy among the amino acids, they can be recoded to increase the chances of accumulating stop mutations during viral replication in viruses with high mutation rates. This approach involves minimal changes to the viral genetic composition and was previously found to rapidly attenuate influenza and coxsackie viruses in vitro and in vivo [21]. An objective of this study was to assess whether the approach of destabilizing serine and leucine codons could be applied to DNA viruses with high mutation rates, using PCV2 as a model. The exact mechanisms by which ssDNA viruses achieve such high mutation rates are as yet unknown but may involve the use of atypical, low fidelity polymerases for replication and repair. Selection pressure induced by factors such as the serial passage in non-specific hosts [18] or conditions of nucleotide depletion [22,23][22][23] can influence mutation rates for single-stranded viruses. In the case of PCV2, vaccination in the field is often undertaken in seropositive piglets due to maternal antibodies or in sows with a history of previous vaccination. Hence, a secondary objective was to study the effect of immune pressure induced by sub-neutralizing antibody levels on the codon destabilized PCV2 vaccine construct.
2. Virus Culture and Cloning of the sPCV2 Vaccine Construct
A previously described PCV2b infectious clone (accession KR816332) [13] was used as the backbone to develop the sPCV2b vaccine. Briefly, the PCV2b genome was amplified from the lung and liver tissues of piglets with clinical signs, using overlapping primers containing a single cutting Sac II enzyme site within the viral genome [13]. To enable the serological monitoring of vaccine compliance in the field, a foreign epitope tag was inserted in the 3′ end of the open reading frame 2 (ORF2), encoding the capsid protein as an independent transcriptional unit. As previously described [24], the epitope tag was derived from an immunogenic peptide located in the surface antigen-1 related sequence 2 (SRS2) protein (AAD04844.1) of Neospora caninum, an Apicomplexan parasite that does not infect pigs. Therefore, antibodies generated against the tag would be detected only in vaccinated pigs. To destabilize the serine and leucine codons, the ORF2 gene sequence between 57 and 609 bps, flanked by BstB1 to MscI enzymes, was commercially synthesized. All serine and leucine codons within the selected sequence were modified as listed in Table 1 to increase the probability that mutations at the 2nd or 3rd positions of the codon that are accrued during vaccine viral replication in the host could result in a stop codon and suicidal replication (Figure 1, Table 1). The synthesized fragment was shuttled into the infectious clone by restriction digestion. The construct is referred to as sPCV2b-Vac in the manuscript.
Figure 1. Map of the sPCV2-Vac construct: The PCV2b backbone is depicted as a circle, with the two major open reading frames indicated by counterclockwise block arrows. The transcriptional unit for the SRS2 epitope tag is shown at the 5′ end of ORF2, which encodes the capsid protein. The location of the modified serine and leucine codons is indicated and flanked by the enzymes used for insertion into the ORF2.
3. Vaccination of Pigs with sPCV2-Vac Protects against Replication of the Heterologous Challenge Virus
Prior to the study, all pigs were screened and confirmed to be PCV2-negative. The vaccine virus was not detected by qPCR at 14- or 28-days post-vaccination (DPV) in the pigs administered sPCV2-Vac, indicating that the codon destabilized PCV2 virus was attenuated in pigs. Following challenge with a heterologous PCV2d virus, viral replication in the unvaccinated pigs followed a pattern typical for PCV2 infection, with virus titers showing an increasing trend during the 21-day post-challenge period. In contrast, the PCV2 challenge virus was not detected in pigs administered the sPCV2-Vac or the commercial control vaccine at DPC 9. At DPC 21, one pig in the sPCV2-Vac group (3.01 ± 2.62 mean log copy numbers/mL serum) and two pigs in the commercial vaccine group (1.92 ± 2.24 mean log copy numbers/mL serum) were very low PCR positives (Figure 3). There was no significant difference between the vaccine groups, and the live virus could not be isolated from the pig sera.
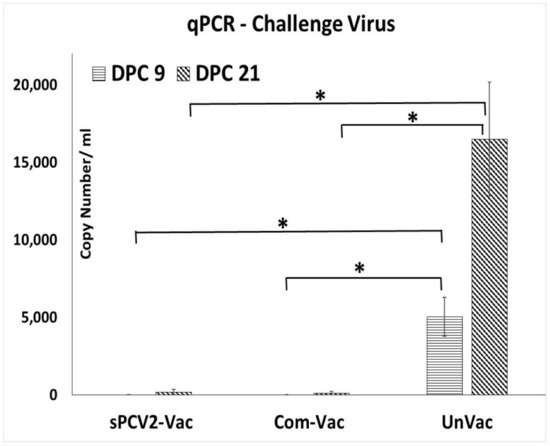

Figure 3. Protection against heterologous challenge viral replication: Challenge viral replication as measured by a PCV2d-specific qPCR. X-axis—treatment groups—horizontal lines—days post-challenge 9, slanted lines—days post-challenge 21, Y-axis—copy numbers/mL serum. Error bars represent the standard error of the mean. * Significantly different from the unvaccinated control group by the Student t-test at p ≤ 0.05 (horizontal bar).
References
- Saiz, J.C. Vaccines against RNA Viruses. Vaccines 2020, 8, 479.
- Stephenson, J.R. Rational design of vaccines against enveloped RNA viruses. Vaccine 1985, 3, 69–72.
- Singh, G.; Singh, P.; Pillatzki, A.; Nelson, E.; Webb, B.; Dillberger-Lawson, S.; Ramamoorthy, S. A Minimally Replicative Vaccine Protects Vaccinated Piglets Against Challenge with the Porcine Epidemic Diarrhea Virus. Front. Vet. Sci. 2019, 6, 347.
- Ramamoorthy, S.; Meng, X.J. Porcine circoviruses: A minuscule yet mammoth paradox. Anim. Health Res. Rev. 2009, 10, 1–20.
- Afghah, Z.; Webb, B.; Meng, X.J.; Ramamoorthy, S. Ten years of PCV2 vaccines and vaccination: Is eradication a possibility? Vet. Microbiol. 2017, 206, 21–28.
- Ssemadaali, M.A.; Ilha, M.; Ramamoorthy, S. Genetic diversity of porcine circovirus type 2 and implications for detection and control. Res. Vet. Sci. 2015, 103, 179–186.
- Franzo, G.; Tucciarone, C.M.; Cecchinato, M.; Drigo, M. Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: A large scale epidemiological study. Sci. Rep. 2016, 6, 39458.
- Gandon, S.; Day, T. Evidences of parasite evolution after vaccination. Vaccine 2008, 26 (Suppl. 3), C4–C7.
- Truyen, U. Evolution of canine parvovirus—A need for new vaccines? Vet. Microbiol. 2006, 117, 9–13.
- Hsu, H.Y.; Chang, M.H.; Ni, Y.H.; Chiang, C.L.; Wu, J.F.; Chen, H.L.; Chen, P.J.; Chen, D.S. Chronologic changes in serum hepatitis B virus DNA, genotypes, surface antigen mutants and reverse transcriptase mutants during 25-year nationwide immunization in Taiwan. J. Viral Hepat. 2017, 24, 645–653.
- Yun, S.I.; Lee, Y.M. Japanese encephalitis: The virus and vaccines. Hum. Vaccines Immunother. 2014, 10, 263–279.
- Padhi, A.; Parcells, M.S. Positive Selection Drives Rapid Evolution of the meq Oncogene of Marek’s Disease Virus. PLoS ONE 2016, 11, e0162180.
- Constans, M.; Ssemadaali, M.; Kolyvushko, O.; Ramamoorthy, S. Antigenic Determinants of Possible Vaccine Escape by Porcine Circovirus Subtype 2b Viruses. Bioinform. Biol. Insights 2015, 9, 1–12.
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821.
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061.
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276.
- Zhao, L.; Rosario, K.; Breitbart, M.; Duffy, S. Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses with Small Genomes and a Diverse Host Range. Adv. Virus Res. 2019, 103, 71–133.
- Allison, A.B.; Kohler, D.J.; Ortega, A.; Hoover, E.A.; Grove, D.M.; Holmes, E.C.; Parrish, C.R. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog. 2014, 10, e1004475.
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42.
- Groenke, N.; Trimpert, J.; Merz, S.; Conradie, A.M.; Wyler, E.; Zhang, H.; Hazapis, O.G.; Rausch, S.; Landthaler, M.; Osterrieder, N.; et al. Mechanism of Virus Attenuation by Codon Pair Deoptimization. Cell Rep. 2020, 31, 107586.
- Moratorio, G.; Henningsson, R.; Barbezange, C.; Carrau, L.; Borderia, A.V.; Blanc, H.; Beaucourt, S.; Poirier, E.Z.; Vallet, T.; Boussier, J.; et al. Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat. Microbiol. 2017, 2, 17088.
- Pereira-Gomez, M.; Sanjuan, R. Delayed lysis confers resistance to the nucleoside analogue 5-fluorouracil and alleviates mutation accumulation in the single-stranded DNA bacteriophage varphiX174. J. Virol. 2014, 88, 5042–5049.
- Stapleford, K.A.; Rozen-Gagnon, K.; Das, P.K.; Saul, S.; Poirier, E.Z.; Blanc, H.; Vidalain, P.O.; Merits, A.; Vignuzzi, M. Viral Polymerase-Helicase Complexes Regulate Replication Fidelity to Overcome Intracellular Nucleotide Depletion. J. Virol. 2015, 89, 11233–11244.
- Rakibuzzaman, A.; Kolyvushko, O.; Singh, G.; Nara, P.; Pineyro, P.; Leclerc, E.; Pillatzki, A.; Ramamoorthy, S. Targeted Alteration of Antibody-Based Immunodominance Enhances the Heterosubtypic Immunity of an Experimental PCV2 Vaccine. Vaccines 2020, 8, 506.
More
