Myeloid neoplasms encompass a very heterogeneous family of diseases characterized by the failure of the molecular mechanisms that ensure a balanced equilibrium between hematopoietic stem cells (HSCs) self-renewal and the proper production of differentiated cells. The origin of the driver mutations leading to preleukemia can be traced back to HSC/progenitor cells. Many properties typical to normal HSCs are exploited by leukemic stem cells (LSCs) to their advantage, leading to the emergence of a clonal population that can eventually progress to leukemia with variable latency and evolution. In fact, different subclones might in turn develop from the original malignant clone through accumulation of additional mutations, increasing their competitive fitness. This process ultimately leads to a complex cancer architecture where a mosaic of cellular clones—each carrying a unique set of mutations—coexists. The repertoire of genes whose mutations contribute to the progression toward leukemogenesis is broad. It encompasses genes involved in different cellular processes, including transcriptional regulation, epigenetics (DNA and histones modifications), DNA damage signaling and repair, chromosome segregation and replication (cohesin complex), RNA splicing, and signal transduction. Among these many players, transcription factors, RNA splicing proteins, and deubiquitinating enzymes are emerging as potential targets for therapeutic intervention.
- myelodysplastic syndromes (MDS)
- acute myeloid leukemia (AML)
- transcription factors
- RNA splicing
- R-loops
- genome integrity
- deubiquitinating enzymes (DUBs)
1. Introduction
Hematopoietic stem cells (HSCs) ensure continuous production of all blood cell types throughout life. By means of a delicate equilibrium between self-renewal, quiescence, and differentiation, HSCs maintain homeostatic conditions and dynamically respond to stress stimuli. The balanced production of the different cell lineages physiologically varies during aging, when the hematopoietic potential is progressively skewed towards the myeloid lineages at the expenses of immune cells [1][2][3]. The accumulation of heritable genetic mutations in individual cells and the kinetics of their selection can lead to myeloid neoplasms, a heterogeneous group of diseases characterized by the dysfunctional production of myeloid cells in the bone marrow. This can manifest in cytopenia and cellular dysplasia, such as in myelodysplastic syndromes (MDSs), in the overproduction of mature clonal myeloid elements, such as in myeloproliferative neoplasms (MPN), or both, as it happens in MDS/MPN, which share different molecular and clinical traits and present both myelodysplastic and proliferative features [4]. Myeloid neoplasms entail a high risk of developing into acute myeloid leukemia (secondary AML or s-AML). AML can also develop as a late complication in patients after leukemogenic therapies (therapy-related AML or t-AML), or without clinical history of prior MDS or known exposure to potentially leukemogenic agents (de novo AML) [5]. Driver mutations leading to a preleukemia condition originate in HSC or in hematopoietic stem-progenitor cells (HSPCs). The preleukemic clones can have a variable latency and, in some cases, can persist for years before further mutations trigger their leukemic evolution [4][6].
MDS has an estimated crude incidence of 4 to 5 cases per 100,000 persons per year. Although MDS occurs at all ages, the incidence is higher in elderly individuals, with a median age of 70 years old at diagnosis [4]. Careful evaluation of the individual patient prognostic risk, genetics, and age guides the clinical decision-making process [4]. Although current drugs can temporarily modulate myelodysplastic hematopoiesis, they fail in eradicating the disease [4][7].
Failure of current therapies in eradicating MDS/AML is attributed to the persistence of leukemic stem cells (LSCs) upon treatment and to the emergence of resistant subclones [8]. Notably, recent deep-sequencing studies revealed the possibility that relapse from chemotherapy can occur not only from LSCs that are resistant to the treatment but also from pre-leukemic mutated but not transformed HSCs, which are already present in the patient at diagnosis or during clinical remission, and which can acquire additional mutations, further highlighting the complexity and heterogeneity of the disease [9][10]. LSCs share functional properties with normal HSCs [11]. Indeed, LSCs are functionally defined as cells capable of self-renewal and of propagating the disease upon transplantation into immunodeficient mice [8]. In addition to self-renewal capacity, LSCs co-opt many survival mechanisms typical to HSCs to their advantage, including genome maintenance processes, epigenetic and stemness transcriptional programs, the pre-mRNA splicing machinery, metabolic properties, interaction with the microenvironment, and inflammatory signals [11][12]. In fact, recurring mutations in regulators of gene expression, including epigenetic proteins, transcription factors (TFs), and the components of the splicing machinery, are found in 70% of AML patients [13][14]. Understanding the molecular mechanisms regulating HSC biology and their dysregulation in pre-leukemic HSCs and in LCSs is therefore critical for understanding the disease and for developing therapies that harness cancer HSPC-specific vulnerabilities and ultimately eradicate the disease.
2. Emerging Therapeutic Targets
2.1. Transcription Factors’ Targeting
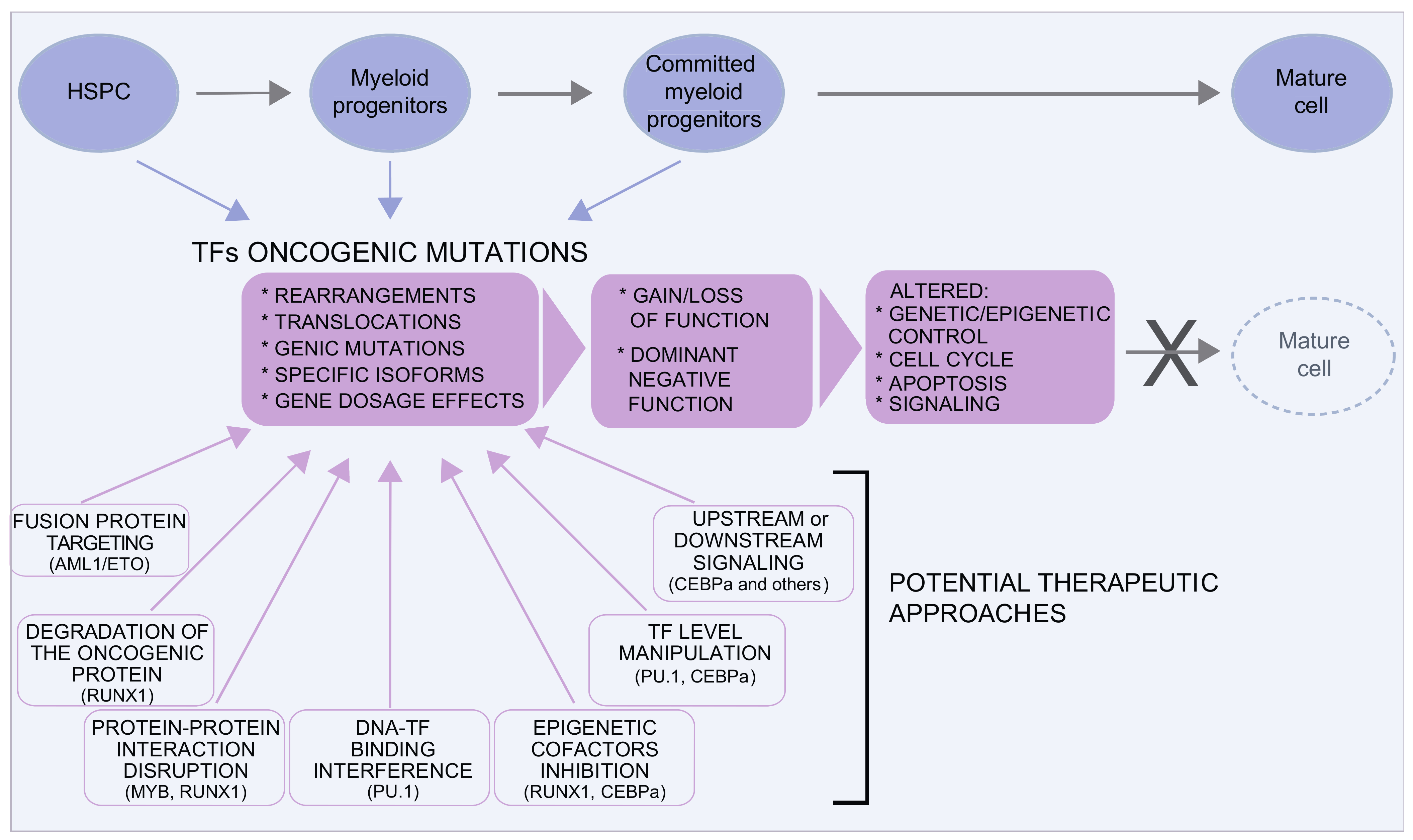 Figure 51. Transcription factors in MDS/AML. Several transcription factors are involved in MDS and AML. Oncogenic mutations are very heterogeneous and can result in gain or loss of function or in dominant negative effects, often with a pleiotropic effect on the control of the equilibrium between proliferation and differentiation. Although traditionally considered “undruggable” factors, in the last years, several approaches have been envisaged to either target the oncogenic protein or to restore the level/function of the TF. Some examples in this direction are discussed in the text and illustrated here.
Figure 51. Transcription factors in MDS/AML. Several transcription factors are involved in MDS and AML. Oncogenic mutations are very heterogeneous and can result in gain or loss of function or in dominant negative effects, often with a pleiotropic effect on the control of the equilibrium between proliferation and differentiation. Although traditionally considered “undruggable” factors, in the last years, several approaches have been envisaged to either target the oncogenic protein or to restore the level/function of the TF. Some examples in this direction are discussed in the text and illustrated here.2.2. Targeting the Spliceosome
2.3. Are DUBs Possible Therapeutic Targets?
3. Conclusions
References
- Morrison, S.J.; Spradling, A.C. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 2008, 132, 598–611.
- De Haan, G.; Lazare, S.S. Aging of hematopoietic stem cells. Blood 2018, 131, 479–487.
- Laurenti, E.; Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553, 418–426.
- Cazzola, M. Myelodysplastic Syndromes. N. Engl. J. Med. 2020, 383, 1358–1374.
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376.
- Sperling, A.S.; Gibson, C.J.; Ebert, B.L. The genetics of myelodysplastic syndrome: From clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer 2017, 17, 5–19.
- Hellstrom-Lindberg, E.; Tobiasson, M.; Greenberg, P. Myelodysplastic syndromes: Moving towards personalized management. Haematologica 2020, 105, 1765–1779.
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737.
- Corces-Zimmerman, M.R.; Hong, W.J.; Weissman, I.L.; Medeiros, B.C.; Majeti, R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA 2014, 111, 2548–2553.
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014, 506, 328–333.
- Yamashita, M.; Dellorusso, P.V.; Olson, O.C.; Passegue, E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat. Rev. Cancer 2020, 20, 365–382.
- Pollyea, D.A.; Jordan, C.T. Therapeutic targeting of acute myeloid leukemia stem cells. Blood 2017, 129, 1627–1635.
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627.
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562, 526–531.
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Chai, J.R.; Lu, J.X.; Zhoa, L.; Gu, L.J.; Wang, Z.Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72, 567–572.
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S.; Ferrara, F.; Fazi, P.; Cicconi, L.; Di Bona, E.; et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013, 369, 111–121.
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474.
- Webb, T.R.; Joyner, A.S.; Potter, P.M. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov. Today 2013, 18, 43–49.
- Yokoi, A.; Kotake, Y.; Takahashi, K.; Kadowaki, T.; Matsumoto, Y.; Minoshima, Y.; Sugi, N.H.; Sagane, K.; Hamaguchi, M.; Iwata, M.; et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011, 278, 4870–4880.
- Teng, T.; Tsai, J.H.C.; Puyang, X.; Seiler, M.; Peng, S.; Prajapati, S.; Aird, D.; Buonamici, S.; Caleb, B.; Chan, B.; et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A–SF3b complex. Nat. Commun. 2017, 8, 15522.
- Cretu, C.; Schmitzová, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Lührmann, R.; Pena, V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 2016, 64, 307–319.
- Finci, L.I.; Zhang, X.; Huang, X.; Zhou, Q.; Tsai, J.; Teng, T.; Agrawal, A.; Chan, B.; Irwin, S.; Karr, C.; et al. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 2018, 32, 309–320.
- Shirai, C.L.; White, B.S.; Tripathi, M.; Tapia, R.; Ley, J.N.; Ndonwi, M.; Kim, S.; Shao, J.; Carver, A.; Saez, B.; et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 2017, 8, 14060.
- Fan, L.; Lagisetti, C.; Edwards, C.C.; Webb, T.R.; Potter, P.M. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 2011, 6, 582–589.
- Convertini, P.; Shen, M.; Potter, P.M.; Palacios, G.; Lagisetti, C.; de la Grange, P.; Horbinski, C.; Fondufe-Mittendorf, Y.N.; Webb, T.R.; Stamm, S. Sudemycin E influences alternative splicing and changes chromatin modifications. Nucleic Acids Res. 2014, 42, 4947–4961.
- Shirai, C.L.; Tripathi, M.; Ley, J.N.; Ndonwi, M.; White, B.S.; Tapia, R.; Saez, B.; Bertino, A.; Shao, J.; Kim, S.; et al. Preclinical Activity of Splicing Modulators in U2AF1 Mutant MDS/AML. Blood 2015, 126, 1653.
- Seiler, M.; Peng, S.; Agrawal, A.A.; Palacino, J.; Teng, T.; Zhu, P.; Smith, P.G.; Buonamici, S.; Yu, L. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep. 2018, 23, 282–296.
- Steensma, D.P.; Wermke, M.; Klimek, V.M.; Greenberg, P.L.; Font, P.; Komrokji, R.S.; Yang, J.; Brunner, A.M.; Carraway, H.E.; Ades, L.; et al. Results of a Clinical Trial of H3B-8800, a Splicing Modulator, in Patients with Myelodysplastic Syndromes (MDS), Acute Myeloid Leukemia (AML) or Chronic Myelomonocytic Leukemia (CMML). Blood 2019, 134, 673.
- Steensma, D.P.; Wermke, M.; Klimek, V.M.; Greenberg, P.L.; Font, P.; Komrokji, R.S.; Yang, J.; Brunner, A.M.; Carraway, H.E.; Ades, L.; et al. Phase I First-in-Human Dose Escalation Study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 2021.
- Jagtap, P.K.A.; Kubelka, T.; Soni, K.; Will, C.L.; Garg, D.; Sippel, C.; Kapp, T.G.; Potukuchi, H.K.; Schorpp, K.; Hadian, K.; et al. Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly. Nat. Commun. 2020, 11, 5621.
- Hong, D.S.; Kurzrock, R.; Naing, A.; Wheler, J.J.; Falchook, G.S.; Schiffman, J.S.; Faulkner, N.; Pilat, M.J.; O’Brien, J.; LoRusso, P. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Investig. New Drugs 2014, 32, 436–444.
- Heideker, J.; Wertz, I.E. DUBs, the regulation of cell identity and disease. Biochem. J. 2015, 467, 191.
- Wertz, I.E.; Wang, X. From Discovery to Bedside: Targeting the Ubiquitin System. Cell Chem. Biol. 2019, 26, 156–177.
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78.
