Fibroblast growth factors (FGFs) comprise a large family of growth factors, regulating diverse biological processes including cell proliferation, migration, and differentiation. Each FGF binds to a set of FGF receptors to initiate certain intracellular signaling molecules. Accumulated evidence suggests that in early development and adult state of vertebrates, FGFs also play exclusive and context dependent roles.
- FGF
- FGFR
- embryonic development
- germ layer formation
- transcription regulation
- embryonic patterning
1. Introduction
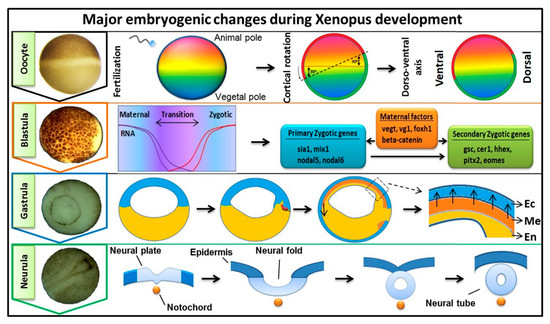
Early embryogenesis in vertebrate embryos involves the irreversible developmental process. As the ovum receives the male haploid genome from a sperm to become a diploid cell, the process of fertilization is started with the fertilized ovum then being referred to as a zygote. The zygote goes through several key developmental stages including mid-blastula transition (MBT), gastrulation (germ layer formation), and neurula, for establishing the overall body axis and generating the anterior CNS and posterior PNS ( Figure 1 ). These are tightly controlled spatiotemporal events led by several signaling pathways and occur in conjunction with maternal or zygotic morphogen gradients throughout the embryos. FGF signaling is known to play an essential role during embryonic development [1[1][2][3],2,3], and in this review, our discussion was focused on involvement of FGF signaling in early embryogenesis. The first discovered FGF ligand, FGF2 (also known as basic FGF/bFGF), was purified from brain tissue in 1975 and was defined for its stimulatory activity in fibroblasts [4]. Since then, a total of 22 FGF members have been identified in humans and similar numbers in vertebrates. Except for the intracellular FGF11 subfamily, these interact with particular FGF receptors (FGFRs) to activate intracellular effector proteins. FGF/FGFR signaling regulates a plethora of cellular processes mediated by activation/modification of cytosolic effectors and followed by transcriptional regulation of target genes. Dysregulation of FGF signaling has been reported to promote several human diseases and disorders, and their severity can vary based on active ligands/receptors and tissues involved [5]. Several lines of evidence also indicate a crucial role for FGF signaling in early embryogenesis for germ layer formation [1,2][1][2] and organogenesis [6]. During primordial germ layer specification, FGFs modulate fate determination as autocrine and paracrine signaling agents. Alterations in tightly regulated FGF expression patterns, as with altered FGFR splicing or mutation and changes in spatiotemporal FGF-FGFR interactions, may result in flawed and defective development for multiple congenital diseases and the onset of various cancer types [5,7,8][5][7][8]. In FGF/FGFR genes, genetic mutations that lead to several congenital diseases have been described reviewed elsewhere [9]. In this review, we summarized the information and our understanding of the functional role(s) of FGFs in early embryonic germ layer specification and axis formation during embryogenesis.
2. FGF and FGFR Families and Signal Transduction
The FGFs are a large family of growth factors consisting of 22 members in humans and mice, and 19 members identified in Xenopus ( Table 1 ). In this family, there are seven subfamilies described in vertebrates, namely FGF1, FGF4, FGF7, FGF8, FGF9, FGF11, and FGF19 (reviewed [10,11][10][11]). Subfamily members have high similarity in their amino acids sequences. Except for the intracellular FGF11 subfamily, extracellular secretion of a given FGF is required for its signaling and function. Based on their secretion profile, FGF members can also be placed into two groups. The first are those whose secretion takes place through a classic endoplasmic reticulum-Golgi secretion pathway in the cells as these FGFs contain N -terminal hydrophobic peptides and they include FGF3, 4, 5, 6, 7, 8, 10, 17, 18, 19, 21, and 23 [10]. The second group of FGFs are those that do not contain N -terminal hydrophobic peptides and are endoplasmic reticulum-Golgi-independent for their secretion. These include FGF1, 2, 9, 16, and 20 [10,11,12][10][11][12]. As an exception, FGF22 remains attached to the cell surface by its N -terminal signal peptide rather than being secreted (reviewed [10]). All members of FGF11 subfamily ( Table 1 ) are known as non-secretory FGFs and are strictly intracellular proteins. Even though these FGFs share structural homologies with other secreted FGFs, they do not share any functional similarities [13,14][13][14]. As intracellular entities, FGF11 subfamily members have been documented as being components of certain protein kinase-mediated signaling pathways and they also interact with membrane channels to regulate cell fate [15,16][15][16].
| Human | Mouse | Xenopus | ||||
|---|---|---|---|---|---|---|
| Subfamily | Members (Other Name) |
Ref. | Members (Other Name) |
Ref. | Members (Other Name) |
Ref. |
| FGF1 | FGF1 (aFGF) | [17] | FGF1 (FGFa) | [18] | xFGF1 | [19] |
| FGF2 (bFGF/FGF-β) | [4] | FGF2 | [20] | xFGF2 (bFGF) | [21] | |
| FGF4 | FGF4 (eFGF) | [22] | FGF4 (KFGF) | [23] | xFGF4 (eFGF, fgf4-a, fgf4-b) | [24] |
| FGF5 | [25] | FGF5 | [26] | xFGF5 | [19][27] | |
| FGF6 (HST2, HSTF2) | [28] | FGF6 (HSTF2) | [29] | xFGF6 | [19][27] | |
| FGF7 | FGF3 | [30] | FGF3 (Int-2) | [31] | xFGF3 (INT-2, FGF3A) | [32] |
| FGF7 (KGF) | [33] | FGF7 (KGF) | [34] | * | ||
| FGF10 | [35] | FGF10 | [36] | xFGF10 | [37] | |
| FGF22 (UNQ2500/PRO5800) | [38] | FGF22 | [38] | xFGF22 | [27] | |
| FGF8 | FGF8 (AIGF) | [39] | FGF8 (AIGF) | [40] | xFGF8 (FGF8a, FGF8b) | [41] |
| FGF17 (UNQ161/PRO187) | [42] | FGF17 | [42] | * | ||
| FGF18 (UNQ420/PRO856) | [43] | FGF18 | [43] | * | ||
| FGF9 | FGF9 | [44] | FGF9 | [45] | xFGF9 (GAF, HBGF9) | [46] |
| FGF16 | [47] | FGF16 | [48] | xFGF16 | [27] | |
| FGF20 | [49] | FGF20 | [50] | xFGF20 | [51] | |
| FGF11 | FGF11 (FHF3) | [52] | FGF11 (FHF3) | [52] | xFGF11 | [27] |
| FGF12 (FGF12B, FHF1) | [52] | FGF12 (FHF1) | [53] | xFGF12 | [54] | |
| FGF13 (FHF2) | [55] | FGF13 (FHF2) | [53] | xFGF13 | [56] | |
| FGF14 (FHF4) | [57] | FGF14 (FHF4) | [58] | xFGF14 (FHF4) | [19] | |
| FGF19 | * | FGF15 | [59] | * | ||
| FGF19 (UNQ334/PRO533) | [60] | * | xFGF19 | [27] | ||
| FGF21 (UNQ3115/PRO10196) | [61] | FGF21 | [61] | * | ||
| FGF23 (HYPF, UNQ3027/PRO9828) | [62] | FGF23 | [62] | xFGF23 (fgf23.1, FGF23.2) | [27] | |
| FGF receptor family | ||||||
| FGFR1 | [17] | FGFR1 | [63] | xFGFR1 | [64] | |
| FGFR2 | [65] | FGFR2 | [66] | xFGFR2 | [67] | |
| FGFR3 | [68] | FGFR3 | [69] | xFGFR3 | [70] | |
| FGFR4 | [71] | FGFR4 | [72] | xFGFR4 | [73] | |
FGFs interact with specific FGF receptors to initiate intracellular signaling and there are four cell membrane tyrosine kinase FGF receptors, FGFR1, 2, 3 and 4, which are members of the larger receptor tyrosine kinase (RTK) group [17,18][74][75]. Each FGFR is a single-pass transmembrane (TM) protein that includes an N -terminal extracellular ligand FGF binding domain and a C -terminal intracellular tyrosine kinase domain. The extracellular domain contains 3 immunoglobulin-like subdomains (D1, D2, and D3 domains) [19][76]. There is an also an acidic box between D1 and D2 domains. The D2 and D3 domains facilitate FGF binding [19][76]. Heparan sulfate (HS) is a coreceptor for FGF binding to an FGFR, and it is essential for FGF binding and signaling. HS is one of the abundant polysaccharides found in the extracellular matrix of mammalian cells [20][77], and it interacts with the cationic patch found in both FGF and D2 subdomain of FGFR [20][77]. Ligand binding induces a conformational change in FGFR, leading to its dimerization and activation of the its intracellular kinase [21][78]. For FGFR1, FGFR2, and FGFR3, two standard isoforms (b-and c-isoforms) are generated by splicing [22][79]. The splicing variants have altered ligand affinity for various FGFs, except for FGF1, acknowledged as a universal ligand and being able to interact with both FGFR isoforms [17,20][74][77].
In this section, we briefly summarized the activation modes of FGFR associated cytosolic effectors and linked components that act as intermediates. Several partner receptor proteins may be associated with the cytosolic domain of an FGFR, such as cell adhesion molecules (CAMs) and G-protein coupled receptors (GPRCs) [23][80]. Signal-induced activation of FGFR typically activates multiple cytosolic signaling pathways. An FGFR is mainly associated with its intracellular signaling intermediates, including phospholipase C (PLCγ), FRS1, FRS2/FRS2α, and FRS3/FRS2β (reviewed [24][81]). FRS2 recruits the adaptor GRB2 (growth factor receptor-bound 2) [25][82], and once GRB2 is bound to the functional domain of the FRS2, it can interact with either SOS or GAB1 and form a complex [26][83]. Upon FGF ligand binding to FGFR and heparan sulfate, multiple cytosolic events occur; these are mostly activational phosphorylations. Once the FGFR complex is activated, GRB2/SOS exchanges the GDP to GTP for Ras; GTP-Ras in turn activates and stimulates Raf (also known as MAPK kinase kinase) as part of the Ras/MAPK pathway (for a detailed mechanism, refer to reviews [27,28,29,30][84][85][86][87]). Similarly, GRB2 switches on PI3K/Akt signaling cascade, as activation of PLCγ/PKC and JAK/STAT pathways are also directly linked to FGFR activation [27,28,29,30][84][85][86][87]. These pathways generate the signals leading to targeted transcription factors regulating transcription of their target genes.
Known FGFs and FGFRs in human, mice, and Xenopus.
3. FGF Signaling in Embryonic Germ Layer Formation
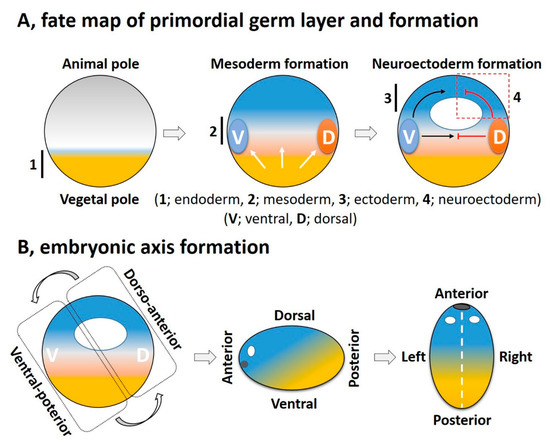
Cellular diversification is an essential process in generating a complex, multicellular organism. This process begins with gastrulation as the three germ layers of endoderm, mesoderm, and ectoderm are specified. For an amphibian embryo, the first embryonic germ to specify is the endoderm from the vegetal pole. The formation of mesoderm, the second germ layer, is the result of active mesoderm inducing signals, originating from the vegetal region or the prospective endoderm, which triggers the mesoderm specification. During the mesoderm specification, generally two distinct signaling centers, namely the ventral and dorsal signaling centers, are established in the marginal region; these are based on presence and activation of maternal or zygotic factors at ventral and dorsal half of the embryo ( Figure 2 A). Once the ventral and dorsal signaling centers are established, they both collectively drive further germ layer specification. The dorsal centers (also called the dorsal mesoderm, dorsal organizer, and organizer) produce dorsal mesoderm and promote neuroectoderm formation by inhibiting ventral signaling. On the opposite end, the ventral center provides the ventral mesoderm and the ectoderm (for epidermis formation) ( Figure 2 A). In this section, we try to summarize primordial germ layer formation and the active role of FGF signaling in this process.
The active molecules with mesoderm inductive activity from the vegetal hemisphere include members of Xenopus nodal-related factors (Xnr1, 2, 4, 5, and 6) and TGFβ family (activin βB), now known as endogenous mesoderm inducing factors in various animal models, including Xenopus, zebrafish, and mouse [106,107,108,109][88][89][90][91] and the signaling pathways utilized by these signals are often due to activin ligands [106,107,108,109][88][89][90][91]. This mesoderm inducing effect by activin treatment in metazoan embryogenesis is now widely accepted. The ectopic expression of activin/Nodal downstream intracellular effectors Smad2/3 activate xbra , chordin (chrd) , noggin (nog) , and goosecoid (gsc) expression [110][92]. Activin-mediated mesoderm induction critically depends on FGF signaling, as demonstrated when DNFR led to complete loss of mesodermal genes expression due to activin [111][93]. In comparison, ectopic expression of Smad2 significantly elevates expression of various FGFs and FGFRs including fgf3 , fgf8 , fgf20 , fgfr1 , and fgfr2 [92][94]. Supporting the role of FGFR signaling in mesoderm induction is also observed with the expression profile of several FGF ligands in early mesoderm of vertebrate embryos. Fgf3 expression is primarily in the ring around blastopore lip (mesoderm) of Xenopus gastrula, which then resolves into neural or brain tissues at a later stage of development [112][95]. Similarly, fgf4 , fgf8 , and fgf20 are largely expressed in early mesoderm or late mesodermal lineages [113,114,115,116][96][97][98][99]. However, integration of FGF and activin/nodal signaling in mesoderm specification is a potential subject for a future investigation.
Brachyury (Bra/Xbra) is a T-box transcription factor and signature mesodermal (also known as a pan-mesodermal marker) factor, and is actively involved in gene regulation process required for mesoderm induction and differentiation [117,118][100][101]. Similarly, Eomesodermin (Eomes) is another T-box transcription factor, an important mesodermal factor crucial for mesoderm induction and differentiation [119][102]. Both proteins are necessary for mesoderm induction; however, Xbra is also involved in expression of ventral/posterior specific genes (e.g., ventx1.1 and xhox3 ) [117,120,121,122][100][103][104][105]. Xbra is indispensable for mesoderm formation and maintenance since knockdown of Xbra converts its mesodermalizing character to a neuralizing one [123,124][106][107]. Eomes has been reported to induce dorsal/anterior mesodermal ( gsc , chrd , and nog ) genes in Xenopus embryos [119][102]. Xbra early expression was reported to depend on FGF signaling as embryos treated with SU5402, showed no xbra expression [114][97]. In turn, FGF2 and FGF4 have been reported to induce xbra transcription in animal explants of Xenopus embryos, wherein Xbra makes an autocatalytic regulatory loop to maintain the Fgf4 expression [123,125][106][108]. Xbra can maintain the expression fgf4 , but its activation is most likely thought to depend on Nodal signaling; this is similar to fgf8 activation largely depending on nodal/activin signaling [114][97]. FGF8, have two alternative splice variants (protein isoforms), namely FGF8a and FGF8b, and these show different inductive features. FGF8a has been reported to have mostly neural inducing activity with little mesoderm inducing capability, whereas FGF8b is important for mesoderm induction and differentiation [55][41].
A spatiotemporal expression of FGF ligands and receptors is seen in Xenopus embryos, suggesting diverse expression patterns among ligands. During gastrulation, fgf1 , fgf2 , fgf4 , fgf8 , fgf20 , and fgf22 are highly expressed, implying a requirement for mesoderm specification and extension [33][19]. Several transcription factors have been reported to incorporate FGF signaling in mesoderm specification and embryonic patterning. As an example, mef2d is transcriptionally activated by FGF4 and FGF8 in the marginal region of Xenopus embryos, and from the vegetal region, nodal (Xnr5 and 6) also induces mef2d expression. Once Mef2d protein is made, it can form a positive expression feedback loop with FGF4 and FGF8, leading to the expression of mesodermal genes like xbra , chrd , gsc , and nog [133][109]. A similar example of interaction with FGF signaling is for Egr1, which has been reported as a downstream target of FGF signaling and plays an important role in embryonic development. The Egr1 activates the transcription of myod , and represses xbra transcription [134][110]. Collectively, the presented evidence suggests that FGF signaling may interact with several other signaling pathways (e.g., activin, nodal, BMP, and RAR) during mesodermal differentiation in a context-dependent manner (for example in the ventral and dorsal mesoderm). However, the details for the different ligands, their receptors, and their mechanistic integration with other key signaling entities remain to be fully elucidated.
4. Regulation of FGF Signaling
Several agents have been discovered or experimentally developed that are relatively specific in modulating FGF signaling, and chemically, these include proteins and small molecules. Based on their activities, these modulators may be placed into three groups: The first includes the endogenous activator(s) as those agents having a positive or compensatory activity relative to FGF signaling. Second are the endogenous factors showing inhibitory or negative activity towards FGF signaling. Third includes the group of relatively small molecular-weight compounds, chemically synthesized to act as inhibitors and with use intended mostly for therapeutic or research applications. Prominently known endogenous activators/inhibitors of the FGF pathway were chosen to be briefly discussed.
A positive feedback regulatory mechanism plays an important role in embryonic and post embryonic development in vertebrates. In this process, several modulators may act as co-activators, interacting with a specific domain of an FGF or an FGFR; this interaction can in turn significantly accelerate or amplify FGF signaling. Notably, heparan sulfate proteoglycans (HSPGs) and glypicans are essential extracellular key modulators, which interact with a variety of growth factors including various FGFs and morphogens and their receptors [197][111]. These interactions can be positive as well as negative on target signaling with fine control being exerted in a spatiotemporal manner. Target signaling may include that for Wnt, Shh, and FGF pathways. Currently, several FGF positive modulators have been identified that include anosmin-1 (An1), Sef1, L1CAM, and FLRT3, and can alter FGF signaling at multiple levels (for more details, we refer the reader to the selected articles [197,198,199,200][111][112][113][114]) ( Table 2 ). Certain small molecules have also been reported to induce FGF signaling. For example, 8-hydroxyquinoline sulfate and pyrithione zinc increases the expression of FGF target genes in zebrafish embryos [199][113]. However, as these reports provide a thoughtful explanation and clues to the regulatory mechanisms in FGF signaling, detailed mechanisms on the control of FGF signaling in development remain to be fully understood.
| Modulators | Target | Effect on FGF Signaling |
|---|---|---|
| Anosmin-1 | Extracellular domain of FGFR | Positive |
| L1CAM | Extracellular domain of FGFR | Positive |
| HSPGs | FGFs or extracellular domain of FGFR | Positive |
| FLRT3 | Intracellular domain of FGFR | Positive |
| Sprouty | Grb/Raf | Negative |
| Sef’s/Spred/Pyst1/Dusp’s | Modification of several kinases activity like (MAPK, MEK, ERK, and PKB/Akt) | Negative |
A negative feedback mechanism is an essential regulatory strategy for nearly all signaling pathways and this is particularly important during embryonic development. FGF signaling has demonstrated as having an active functional role at multiple steps across embryonic development. To date, a group of endogenous proteins have been identified that show antagonist activity to FGF signals and are generally associated with the negative feedback loop in downregulating FGF signal transduction. One is Sprouty, an intracellular protein that interacts with Drk and Gap1, being components of Ras/MAPK pathway, and blocks their interaction [197,198][111][112]. Another is Sef, with a similar expression to various fgf genes, which is capable of interacting with the cytosolic domain of FGFR and blocking the intracellular effectors interacting with FGFR. An ectopic gain of function of sef , for example, has been shown to reduce phosphorylation of Raf1 and MEK1/2 [199][113]. Other agents that have been reported are Spred [200][114], Pyst1/Mkp3 [197][111], Dusp1 [92][94], and FRS2α/β [201][115], all endogenous FGF inhibitors/modulators. All of the above mentioned blockers of FGF signaling may have separate or overlapping targets (see Table 2 ) and are generally cytosolic effector proteins.
FGF modulators are essential in early embryogenesis as several studies have reported knockdown of various HSPG genes causing defective development in vertebrates and invertebrates (reviewed [202][116]). The initial reports from a vertebrate model, namely mouse, has the mutation of “lazy mesoderm” ( lzme ) gene where lzme encodes an enzyme required for glycosaminoglycan biosynthesis) causing defective mesoderm and endoderm migration [203][117]. Gene expression profiles demonstrated that FGF target genes expression are severely compromised in lzme mutant embryos, whereas the nodal and Wnt3 pathways remained normal [203][117]. Similar findings have been reported in Xenopus embryos, wherein morpholino-based knockdown of gpc4 (Glypican 4) shows pleiotropic developmental aberrant phenotypes including defected primary axis and forebrain patterning [204][118]. A detailed discussion of FGF modulators, however, is beyond the focus of current review, and we refer the readers to the related articles [202,203,204,205,206,207][116][117][118][119][120][121].
References
- Bottcher, R.T.; Niehrs, C. Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 2005, 26, 63–77.
- Dorey, K.; Amaya, E. FGF signalling: Diverse roles during early vertebrate embryogenesis. Development 2010, 137, 3731–3742.
- Pownall, M.E.; Isaacs, H.V. FGF signalling in vertebrate development. Colloq. Ser. Dev. Biol. 2010, 1, 1–75.
- Florkiewicz, R.Z.; Shibata, F.; Barankiewicz, T.; Baird, A.; Gonzalez, A.M.; Florkiewicz, E.; Shah, N. Basic fibroblast growth factor gene expression. Ann. N. Y. Acad. Sci. 1991, 638, 109–126.
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal. Transduct. Target. Ther. 2020, 5, 181.
- Kato, S.; Sekine, K. FGF-FGFR signaling in vertebrate organogenesis. Cell Mol. Biol. 1999, 45, 631–638.
- Katoh, M.; Nakagama, H. FGF receptors: Cancer biology and therapeutics. Med. Res. Rev. 2014, 34, 280–300.
- Coumoul, X.; Deng, C.X. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res. C Embryo Today 2003, 69, 286–304.
- Moosa, S.; Wollnik, B. Altered FGF signalling in congenital craniofacial and skeletal disorders. Semin. Cell Dev. Biol. 2016, 53, 115–125.
- Itoh, N.; Ornitz, D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004, 20, 563–569.
- Ornitz, D.M.; Itoh, N. Fibroblast growth factors. Genome Biol. 2001, 2, REVIEWS3005.
- Hui, Q.; Jin, Z.; Li, X.; Liu, C.; Wang, X. FGF Family: From Drug Development to Clinical Application. Int. J. Mol. Sci. 2018, 19, 1875.
- Olsen, S.K.; Garbi, M.; Zampieri, N.; Eliseenkova, A.V.; Ornitz, D.M.; Goldfarb, M.; Mohammadi, M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 2003, 278, 34226–34236.
- Schoorlemmer, J.; Goldfarb, M. Fibroblast growth factor homologous factors are intracellular signaling proteins. Curr. Biol. 2001, 11, 793–797.
- Yan, H.; Pablo, J.L.; Pitt, G.S. FGF14 regulates presynaptic Ca2+ channels and synaptic transmission. Cell Rep. 2013, 4, 66–75.
- Wu, Q.F.; Yang, L.; Li, S.; Wang, Q.; Yuan, X.B.; Gao, X.; Bao, L.; Zhang, X. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell 2012, 149, 1549–1564.
- Dionne, C.A.; Crumley, G.; Bellot, F.; Kaplow, J.M.; Searfoss, G.; Ruta, M.; Burgess, W.H.; Jaye, M.; Schlessinger, J. Cloning and expression of two distinct high-affinity receptors cross-reacting with acidic and basic fibroblast growth factors. EMBO J. 1990, 9, 2685–2692.
- Madiai, F.; Hackshaw, K.V.; Chiu, I.M. Cloning and characterization of the mouse Fgf-1 gene. Gene 1996, 179, 231–236.
- Lea, R.; Papalopulu, N.; Amaya, E.; Dorey, K. Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev. Dyn. 2009, 238, 1467–1479.
- Hebert, J.M.; Basilico, C.; Goldfarb, M.; Haub, O.; Martin, G.R. Isolation of cDNAs encoding four mouse FGF family members and characterization of their expression patterns during embryogenesis. Dev. Biol. 1990, 138, 454–463.
- Kimelman, D.; Kirschner, M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell 1987, 51, 869–877.
- Feldman, B.; Poueymirou, W.; Papaioannou, V.E.; DeChiara, T.M.; Goldfarb, M. Requirement of FGF-4 for postimplantation mouse development. Science 1995, 267, 246–249.
- Brookes, S.; Smith, R.; Thurlow, J.; Dickson, C.; Peters, G. The mouse homologue of hst/k-FGF: Sequence, genome organization and location relative to int-2. Nucleic Acids Res. 1989, 17, 4037–4045.
- Isaacs, H.V.; Tannahill, D.; Slack, J.M. Expression of a novel FGF in the Xenopus embryo. A new candidate inducing factor for mesoderm formation and anteroposterior specification. Development 1992, 114, 711–720.
- Zhan, X.; Bates, B.; Hu, X.G.; Goldfarb, M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol. Cell Biol. 1988, 8, 3487–3495.
- Ozawa, K.; Suzuki, S.; Asada, M.; Tomooka, Y.; Li, A.J.; Yoneda, A.; Komi, A.; Imamura, T. An alternatively spliced fibroblast growth factor (FGF)-5 mRNA is abundant in brain and translates into a partial agonist/antagonist for FGF-5 neurotrophic activity. J. Biol. Chem. 1998, 273, 29262–29271.
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343.
- Iida, S.; Yoshida, T.; Naito, K.; Sakamoto, H.; Katoh, O.; Hirohashi, S.; Sato, T.; Onda, M.; Sugimura, T.; Terada, M. Human hst-2 (FGF-6) oncogene: cDNA cloning and characterization. Oncogene 1992, 7, 303–309.
- De Lapeyriere, O.; Rosnet, O.; Benharroch, D.; Raybaud, F.; Marchetto, S.; Planche, J.; Galland, F.; Mattei, M.G.; Copeland, N.G.; Jenkins, N.A.; et al. Structure, chromosome mapping and expression of the murine Fgf-6 gene. Oncogene 1990, 5, 823–831.
- Brookes, S.; Smith, R.; Casey, G.; Dickson, C.; Peters, G. Sequence organization of the human int-2 gene and its expression in teratocarcinoma cells. Oncogene 1989, 4, 429–436.
- Dickson, C.; Acland, P.; Smith, R.; Dixon, M.; Deed, R.; MacAllan, D.; Walther, W.; Fuller-Pace, F.; Kiefer, P.; Peters, G. Characterization of int-2: A member of the fibroblast growth factor family. J. Cell Sci. Suppl. 1990, 13, 87–96.
- Kiefer, P.; Mathieu, M.; Close, M.J.; Peters, G.; Dickson, C. FGF3 from Xenopus laevis. EMBO J. 1993, 12, 4159–4168.
- Finch, P.W.; Rubin, J.S.; Miki, T.; Ron, D.; Aaronson, S.A. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science 1989, 245, 752–755.
- Mason, I.J.; Fuller-Pace, F.; Smith, R.; Dickson, C. FGF-7 (keratinocyte growth factor) expression during mouse development suggests roles in myogenesis, forebrain regionalisation and epithelial-mesenchymal interactions. Mech. Dev. 1994, 45, 15–30.
- Emoto, H.; Tagashira, S.; Mattei, M.G.; Yamasaki, M.; Hashimoto, G.; Katsumata, T.; Negoro, T.; Nakatsuka, M.; Birnbaum, D.; Coulier, F.; et al. Structure and expression of human fibroblast growth factor-10. J. Biol. Chem. 1997, 272, 23191–23194.
- Tagashira, S.; Harada, H.; Katsumata, T.; Itoh, N.; Nakatsuka, M. Cloning of mouse FGF10 and up-regulation of its gene expression during wound healing. Gene 1997, 197, 399–404.
- Yokoyama, H.; Ide, H.; Tamura, K. FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev. Biol. 2001, 233, 72–79.
- Nakatake, Y.; Hoshikawa, M.; Asaki, T.; Kassai, Y.; Itoh, N. Identification of a novel fibroblast growth factor, FGF-22, preferentially expressed in the inner root sheath of the hair follicle. Biochim. Biophys. Acta 2001, 1517, 460–463.
- Tanaka, A.; Miyamoto, K.; Matsuo, H.; Matsumoto, K.; Yoshida, H. Human androgen-induced growth factor in prostate and breast cancer cells: Its molecular cloning and growth properties. FEBS Lett. 1995, 363, 226–230.
- Tanaka, A.; Miyamoto, K.; Minamino, N.; Takeda, M.; Sato, B.; Matsuo, H.; Matsumoto, K. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proc. Natl. Acad. Sci. USA 1992, 89, 8928–8932.
- Fletcher, R.B.; Baker, J.C.; Harland, R.M. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 2006, 133, 1703–1714.
- Hoshikawa, M.; Ohbayashi, N.; Yonamine, A.; Konishi, M.; Ozaki, K.; Fukui, S.; Itoh, N. Structure and expression of a novel fibroblast growth factor, FGF-17, preferentially expressed in the embryonic brain. Biochem. Biophys. Res. Commun. 1998, 244, 187–191.
- Hu, M.C.; Qiu, W.R.; Wang, Y.P.; Hill, D.; Ring, B.D.; Scully, S.; Bolon, B.; DeRose, M.; Luethy, R.; Simonet, W.S.; et al. FGF-18, a novel member of the fibroblast growth factor family, stimulates hepatic and intestinal proliferation. Mol. Cell Biol. 1998, 18, 6063–6074.
- Miyamoto, M.; Naruo, K.; Seko, C.; Matsumoto, S.; Kondo, T.; Kurokawa, T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol. Cell Biol. 1993, 13, 4251–4259.
- Santos-Ocampo, S.; Colvin, J.S.; Chellaiah, A.; Ornitz, D.M. Expression and biological activity of mouse fibroblast growth factor-9. J. Biol. Chem. 1996, 271, 1726–1731.
- Song, J.; Slack, J.M. XFGF-9: A new fibroblast growth factor from Xenopus embryos. Dev. Dyn. 1996, 206, 427–436.
- Miyake, A.; Konishi, M.; Martin, F.H.; Hernday, N.A.; Ozaki, K.; Yamamoto, S.; Mikami, T.; Arakawa, T.; Itoh, N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem. Biophys. Res. Commun. 1998, 243, 148–152.
- Sontag, D.P.; Cattini, P.A. Cloning and bacterial expression of postnatal mouse heart FGF-16. Mol. Cell Biochem. 2003, 242, 65–70.
- Kirikoshi, H.; Sagara, N.; Saitoh, T.; Tanaka, K.; Sekihara, H.; Shiokawa, K.; Katoh, M. Molecular cloning and characterization of human FGF-20 on chromosome 8p21.3-p22. Biochem. Biophys. Res. Commun. 2000, 274, 337–343.
- Hajihosseini, M.K.; Heath, J.K. Expression patterns of fibroblast growth factors-18 and -20 in mouse embryos is suggestive of novel roles in calvarial and limb development. Mech. Dev. 2002, 113, 79–83.
- Koga, C.; Adati, N.; Nakata, K.; Mikoshiba, K.; Furuhata, Y.; Sato, S.; Tei, H.; Sakaki, Y.; Kurokawa, T.; Shiokawa, K.; et al. Characterization of a novel member of the FGF family, XFGF-20, in Xenopus laevis. Biochem. Biophys. Res. Commun. 1999, 261, 756–765.
- Smallwood, P.M.; Munoz-Sanjuan, I.; Tong, P.; Macke, J.P.; Hendry, S.H.; Gilbert, D.J.; Copeland, N.G.; Jenkins, N.A.; Nathans, J. Fibroblast growth factor (FGF) homologous factors: New members of the FGF family implicated in nervous system development. Proc. Natl. Acad. Sci. USA 1996, 93, 9850–9857.
- Hartung, H.; Feldman, B.; Lovec, H.; Coulier, F.; Birnbaum, D.; Goldfarb, M. Murine FGF-12 and FGF-13: Expression in embryonic nervous system, connective tissue and heart. Mech. Dev. 1997, 64, 31–39.
- Fukui, L.; Henry, J.J. FGF signaling is required for lens regeneration in Xenopus laevis. Biol. Bull. 2011, 221, 137–145.
- Gecz, J.; Baker, E.; Donnelly, A.; Ming, J.E.; McDonald-McGinn, D.M.; Spinner, N.B.; Zackai, E.H.; Sutherland, G.R.; Mulley, J.C. Fibroblast growth factor homologous factor 2 (FHF2): Gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum. Genet. 1999, 104, 56–63.
- Nishimoto, S.; Nishida, E. Fibroblast growth factor 13 is essential for neural differentiation in Xenopus early embryonic development. J. Biol. Chem. 2007, 282, 24255–24261.
- Goetz, R.; Dover, K.; Laezza, F.; Shtraizent, N.; Huang, X.; Tchetchik, D.; Eliseenkova, A.V.; Xu, C.F.; Neubert, T.A.; Ornitz, D.M.; et al. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 2009, 284, 17883–17896.
- Yamamoto, S.; Mikami, T.; Konishi, M.; Itoh, N. Stage-specific expression of a novel isoform of mouse FGF-14 (FHF-4) in spermatocytes. Biochim. Biophys. Acta 2000, 1490, 121–124.
- Vergnes, L.; Lee, J.M.; Chin, R.G.; Auwerx, J.; Reue, K. Diet1 functions in the FGF15/19 enterohepatic signaling axis to modulate bile acid and lipid levels. Cell Metab. 2013, 17, 916–928.
- Nishimura, T.; Utsunomiya, Y.; Hoshikawa, M.; Ohuchi, H.; Itoh, N. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim. Biophys. Acta 1999, 1444, 148–151.
- Nishimura, T.; Nakatake, Y.; Konishi, M.; Itoh, N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim. Biophys. Acta 2000, 1492, 203–206.
- Yamashita, T.; Yoshioka, M.; Itoh, N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem. Biophys. Res. Commun. 2000, 277, 494–498.
- Reid, H.H.; Wilks, A.F.; Bernard, O. Two forms of the basic fibroblast growth factor receptor-like mRNA are expressed in the developing mouse brain. Proc. Natl. Acad. Sci. USA 1990, 87, 1596–1600.
- Friesel, R.; Dawid, I.B. cDNA cloning and developmental expression of fibroblast growth factor receptors from Xenopus laevis. Mol. Cell Biol. 1991, 11, 2481–2488.
- Zhang, Y.; Gorry, M.C.; Post, J.C.; Ehrlich, G.D. Genomic organization of the human fibroblast growth factor receptor 2 (FGFR2) gene and comparative analysis of the human FGFR gene family. Gene 1999, 230, 69–79.
- Orr-Urtreger, A.; Bedford, M.T.; Burakova, T.; Arman, E.; Zimmer, Y.; Yayon, A.; Givol, D.; Lonai, P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 1993, 158, 475–486.
- Friesel, R.; Brown, S.A. Spatially restricted expression of fibroblast growth factor receptor-2 during Xenopus development. Development 1992, 116, 1051–1058.
- Keegan, K.; Johnson, D.E.; Williams, L.T.; Hayman, M.J. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc. Natl. Acad. Sci. USA 1991, 88, 1095–1099.
- Chellaiah, A.T.; McEwen, D.G.; Werner, S.; Xu, J.; Ornitz, D.M. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J. Biol. Chem. 1994, 269, 11620–11627.
- Hongo, I.; Kengaku, M.; Okamoto, H. FGF signaling and the anterior neural induction in Xenopus. Dev. Biol. 1999, 216, 561–581.
- Partanen, J.; Makela, T.P.; Eerola, E.; Korhonen, J.; Hirvonen, H.; Claesson-Welsh, L.; Alitalo, K. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991, 10, 1347–1354.
- Stark, K.L.; McMahon, J.A.; McMahon, A.P. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development 1991, 113, 641–651.
- Riou, J.F.; Clavilier, L.; Boucaut, J.C. Early regionalized expression of a novel Xenopus fibroblast growth factor receptor in neuroepithelium. Biochem. Biophys. Res. Commun. 1996, 218, 198–204.
- Ornitz, D.M.; Xu, J.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996, 271, 15292–15297.
- Givol, D.; Yayon, A. Complexity of FGF receptors: Genetic basis for structural diversity and functional specificity. FASEB J. 1992, 6, 3362–3369.
- Farrell, B.; Breeze, A.L. Structure, activation and dysregulation of fibroblast growth factor receptor kinases: Perspectives for clinical targeting. Biochem. Soc. Trans. 2018, 46, 1753–1770.
- Brown, A.; Robinson, C.J.; Gallagher, J.T.; Blundell, T.L. Cooperative heparin-mediated oligomerization of fibroblast growth factor-1 (FGF1) precedes recruitment of FGFR2 to ternary complexes. Biophys. J. 2013, 104, 1720–1730.
- Sarabipour, S.; Hristova, K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016, 7, 10262.
- Werner, S.; Duan, D.S.; de Vries, C.; Peters, K.G.; Johnson, D.E.; Williams, L.T. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol. Cell Biol. 1992, 12, 82–88.
- Latko, M.; Czyrek, A.; Porebska, N.; Kucinska, M.; Otlewski, J.; Zakrzewska, M.; Opalinski, L. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455.
- Zhang, J.; Tang, P.M.K.; Zhou, Y.; Cheng, A.S.L.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic FGF-FGFR Axis in Gastric Carcinogenesis. Cells 2019, 8, 637.
- Zhang, Y.; McKeehan, K.; Lin, Y.; Zhang, J.; Wang, F. Fibroblast growth factor receptor 1 (FGFR1) tyrosine phosphorylation regulates binding of FGFR substrate 2alpha (FRS2alpha) but not FRS2 to the receptor. Mol. Endocrinol. 2008, 22, 167–175.
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180.
- Dey, J.H.; Bianchi, F.; Voshol, J.; Bonenfant, D.; Oakeley, E.J.; Hynes, N.E. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010, 70, 4151–4162.
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129.
- Eswarakumar, V.P.; Lax, I.; Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005, 16, 139–149.
- Tsang, M.; Dawid, I.B. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci. STKE 2004, 2004, pe17.
- Takenaga, M.; Fukumoto, M.; Hori, Y. Regulated Nodal signaling promotes differentiation of the definitive endoderm and mesoderm from ES cells. J. Cell Sci. 2007, 120, 2078–2090.
- Agius, E.; Oelgeschlager, M.; Wessely, O.; Kemp, C.; de Robertis, E.M. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development 2000, 127, 1173–1183.
- Feldman, B.; Gates, M.A.; Egan, E.S.; Dougan, S.T.; Rennebeck, G.; Sirotkin, H.I.; Schier, A.F.; Talbot, W.S. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 1998, 395, 181–185.
- Jones, C.M.; Kuehn, M.R.; Hogan, B.L.; Smith, J.C.; Wright, C.V. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development 1995, 121, 3651–3662.
- Kumar, V.; Umair, Z.; Kumar, S.; Lee, U.; Kim, J. Smad2 and Smad3 differentially modulate chordin transcription via direct binding on the distal elements in gastrula Xenopus embryos. Biochem. Biophys. Res. Commun. 2021, 559, 168–175.
- Cornell, R.A.; Musci, T.J.; Kimelman, D. FGF is a prospective competence factor for early activin-type signals in Xenopus mesoderm induction. Development 1995, 121, 2429–2437.
- Umair, Z.; Kumar, S.; Rafiq, K.; Kumar, V.; Reman, Z.U.; Lee, S.H.; Kim, S.; Lee, J.Y.; Lee, U.; Kim, J. Dusp1 modulates activin/smad2 mediated germ layer specification via FGF signal inhibition in Xenopus embryos. Anim. Cells Syst. 2020, 24, 359–370.
- Tannahill, D.; Isaacs, H.V.; Close, M.J.; Peters, G.; Slack, J.M. Developmental expression of the Xenopus int-2 (FGF-3) gene: Activation by mesodermal and neural induction. Development 1992, 115, 695–702.
- Branney, P.A.; Faas, L.; Steane, S.E.; Pownall, M.E.; Isaacs, H.V. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS ONE 2009, 4, e4951.
- Fletcher, R.B.; Harland, R.M. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev. Dyn. 2008, 237, 1243–1254.
- Christen, B.; Slack, J.M. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev. Biol. 1997, 192, 455–466.
- Isaacs, H.V. New perspectives on the role of the fibroblast growth factor family in amphibian development. Cell Mol. Life Sci. 1997, 53, 350–361.
- Conlon, F.L.; Sedgwick, S.G.; Weston, K.M.; Smith, J.C. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development 1996, 122, 2427–2435.
- Schulte-Merker, S.; Smith, J.C. Mesoderm formation in response to Brachyury requires FGF signalling. Curr. Biol. 1995, 5, 62–67.
- Ryan, K.; Garrett, N.; Mitchell, A.; Gurdon, J.B. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell 1996, 87, 989–1000.
- Kumar, S.; Umair, Z.; Yoon, J.; Lee, U.; Kim, S.C.; Park, J.B.; Lee, J.Y.; Kim, J. Xbra and Smad-1 cooperate to activate the transcription of neural repressor ventx1.1 in Xenopus embryos. Sci. Rep. 2018, 8, 11391.
- Brewster, R.; Mullor, J.L.; Ruiz i Altaba, A. Gli2 functions in FGF signaling during antero-posterior patterning. Development 2000, 127, 4395–4405.
- Kim, J.; Lin, J.J.; Xu, R.H.; Kung, H.F. Mesoderm induction by heterodimeric AP-1 (c-Jun and c-Fos) and its involvement in mesoderm formation through the embryonic fibroblast growth factor/Xbra autocatalytic loop during the early development of Xenopus embryos. J. Biol. Chem. 1998, 273, 1542–1550.
- Yoon, J.; Kim, J.H.; Lee, S.Y.; Kim, S.; Park, J.B.; Lee, J.Y.; Kim, J. PV. 1 induced by FGF-Xbra functions as a repressor of neurogenesis in Xenopus embryos. BMB Rep. 2014, 47, 673–678.
- Rao, Y. Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes Dev. 1994, 8, 939–947.
- Isaacs, H.V.; Pownall, M.E.; Slack, J.M. eFGF regulates Xbra expression during Xenopus gastrulation. EMBO J. 1994, 13, 4469–4481.
- Kolpakova, A.; Katz, S.; Keren, A.; Rojtblat, A.; Bengal, E. Transcriptional regulation of mesoderm genes by MEF2D during early Xenopus development. PLoS ONE 2013, 8, e69693.
- Nentwich, O.; Dingwell, K.S.; Nordheim, A.; Smith, J.C. Downstream of FGF during mesoderm formation in Xenopus: The roles of Elk-1 and Egr-1. Dev. Biol. 2009, 336, 313–326.
- Eblaghie, M.C.; Lunn, J.S.; Dickinson, R.J.; Munsterberg, A.E.; Sanz-Ezquerro, J.J.; Farrell, E.R.; Mathers, J.; Keyse, S.M.; Storey, K.; Tickle, C. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr. Biol. 2003, 13, 1009–1018.
- Casci, T.; Vinos, J.; Freeman, M. Sprouty, an intracellular inhibitor of Ras signaling. Cell 1999, 96, 655–665.
- Kovalenko, D.; Yang, X.; Nadeau, R.J.; Harkins, L.K.; Friesel, R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J. Biol. Chem. 2003, 278, 14087–14091.
- Sivak, J.M.; Petersen, L.F.; Amaya, E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev. Cell 2005, 8, 689–701.
- Lax, I.; Wong, A.; Lamothe, B.; Lee, A.; Frost, A.; Hawes, J.; Schlessinger, J. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell 2002, 10, 709–719.
- Fico, A.; Maina, F.; Dono, R. Fine-tuning of cell signaling by glypicans. Cell Mol. Life Sci. 2011, 68, 923–929.
- Garcia-Garcia, M.J.; Anderson, K.V. Essential role of glycosaminoglycans in Fgf signaling during mouse gastrulation. Cell 2003, 114, 727–737.
- Galli, A.; Roure, A.; Zeller, R.; Dono, R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development 2003, 130, 4919–4929.
- Korsensky, L.; Ron, D. Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Semin. Cell Dev. Biol. 2016, 53, 101–114.
- Saydmohammed, M.; Vollmer, L.L.; Onuoha, E.O.; Vogt, A.; Tsang, M. A high-content screening assay in transgenic zebrafish identifies two novel activators of fgf signaling. Birth Defects Res. C Embryo Today 2011, 93, 281–287.
- Jaeger, I.; Arber, C.; Risner-Janiczek, J.R.; Kuechler, J.; Pritzsche, D.; Chen, I.C.; Naveenan, T.; Ungless, M.A.; Li, M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development 2011, 138, 4363–4374.
