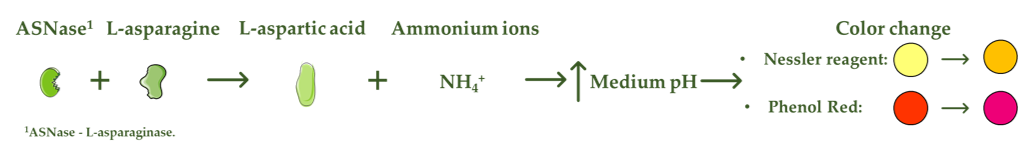
L-asparaginase (ASNase) is an aminohydrolase enzyme widely used in the pharmaceutical and food industries. Although currently its main applications are focused on the treatment of lymphoproliferative disorders such as acute lymphoblastic leukemia (ALL) and acrylamide reduction in starch-rich foods cooked at temperatures above 100 °C, its use as a biosensor in the detection and monitoring of L-asparagine levels is of high relevance. ASNase-based biosensors are a promising and innovative technology, mostly based on colorimetric detection since the mechanism of action of ASNase is the catalysis of the L-asparagine hydrolysis, which releases L-aspartic acid and ammonium ions, promoting a medium pH value change followed by color variation. ASNase biosensing systems prove their potential for L-asparagine monitoring in ALL patients, along with L-asparagine concentration analysis in foods, due to their simplicity and fast response.
- L-asparaginase
- biosensor
- L-asparagine
- monitoring
- ammonia
- pharmaceutical
- food
- industry
- Introduction
Biosensors are analytical systems consisting of an immobilized biological element combined with a suitable transducer to quantify an analyte [1]. Depending on the biosensor purpose, the most common transducers are electrochemical, piezoelectric, optical, thermometric or magnetic [2]. Biosensors usually generate an electronic or optical signal proportional to the particular interaction between the analyte and the immobilized recognition compound. The low equipment costs, high sensitivity and precision and easy operation have led to increased demand for this type of systems compared to traditional analytical methods [3]. The biological component may be constituted by different bio-recognition elements, such as enzymes, bacteria, tissues, antibodies or nucleic acids, etc. Among them, enzyme-based biosensors have been increasingly used due to their recognized high specificity and exceptional biorecognition capabilities [4]. The enzyme immobilization procedure can influence the biosensor stability, specificity, sensitivity and reproducibility. Therefore, the method used to attach the enzyme onto the electrode must guarantee the stability of the active site and the biomolecule activity. The selection of a suitable technique depends on the biological component nature, the transducer type, the analyte characteristics and the operating conditions. The most used enzyme immobilization methods for the design and development of biosensors are classified into physical adsorption, covalent binding and entrapment [5]. Physical adsorption comprises low associated costs and improved enzymatic performance, whereas entrapment (within the framework of a support) allows enzyme preservation and high enzymatic activity levels. However, physical adsorption can lead to nonspecific adsorption and enzyme desorption, while entrapment can lead to mass transfer limitations and enzyme leakage. On the other hand, by applying a covalent bond between the enzyme and the immobilization support, the enzyme leaching is avoided, allowing for the recovery and reuse of the support/enzyme. Nevertheless, in this last option, the enzyme’s active center amino acids should not be involved in covalent bonding to guarantee high levels of enzyme activity, which is defined by the enzyme binding orientation [6,7].
The enzyme L-asparaginase (E.C.3.5.1.1, ASNase) is a chemotherapeutic agent for the treatment of lymphoproliferative disorders, specifically acute lymphoblastic leukemia (ALL), lymphomas and natural killer cell tumors. The tumor-inhibitory characteristics of ASNase were first reported in animal trials in the 1950s [8,9]. ASNase catalyzes the hydrolysis of L-asparagine into L-aspartic acid and ammonia. Since healthy cells can produce L-asparagine internally, whereas leukemic cells depend on this crucial extracellular amino acid for their growth, prolonged deprivation of L-asparagine in blood leads malignant lymphoid cells into apoptosis [10]. L-asparagine concentration varies from 10
to 10
M in healthy blood serum samples and from 10
to 10
M in leukemia blood serum samples [11]. Thus, monitoring L-asparagine depletion in ALL patients is essential to assess the efficacy of ASNase therapy [12]. Full L-asparagine depletion and high ASNase activity are both associated with improved outcomes in ALL patients [13].
ASNase also has an important application in the food industry for acrylamide mitigation from heat-processed foods. Acrylamide is referred to as a Group 2A carcinogen (“probably carcinogenic to humans”) by the International Agency for Research on Cancer (IARC) and by the World Health Organization (WHO) [14,15]. According to the Food and Drug Administration (FDA), the food products with the highest acrylamide concentrations (up to 8440 µg.kg
) are cereals, french fries, potato chips and cookies, whose average daily acrylamide intake varies from 0.03 to 0.05 µg.kg
body weight [6,16]. Nevertheless, the tolerable daily acrylamide intake to avoid carcinogenic risk is 2.6 µg kg
body weight [17]. The pre-treatment of starchy foods with ASNase before cooking transforms L-asparagine into L-aspartic acid, and the Maillard reaction takes place without the contribution of L-asparagine, avoiding the acrylamide formation in the final food product [18]. Since the amino acid L-asparagine is not mainly responsible for the taste and appearance of the processed foods, the desired organoleptic characteristics are preserved [19]. Thus, ASNase is already used to lower the acrylamide dosage in several food products, such as potatoes, bread, french fries, coffee and biscuits [6].
ASNase-based biosensors are a promising and innovative technology that can be used both to detect and monitor the level of L-asparagine in blood serum samples of leukemic patients and in different food samples [6]. The methods currently used to detect L-asparagine are based on spectroscopy and chromatography techniques [20], whereas the biosensor operation mode is mostly based on colorimetric detection. The L-asparagine hydrolysis releases ammonium ions, promoting, consequently, a change in the pH value of the medium followed by a variation in the color [21], being the mode of action of colorimetric ASNase-based biosensors (Figure 1). Although there are few reports on the development of ASNase-based biosensors, biosensing systems prove to be imperative for pharmaceutical/food industrial applications due to their simplicity and fast response, while allowing online L-asparagine monitoring.
. Mode of action of colorimetric ASNase based biosensors.
- Types and Applications of L-Asparaginase-Based Biosensors
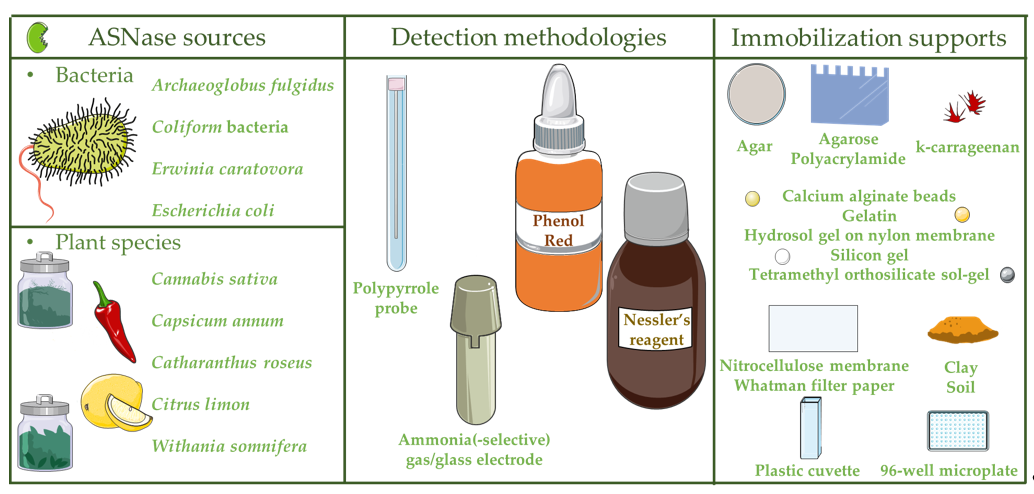
In 1983, the first ASNase-based biosensor was developed for ammonia detection through the combination of an online gas dialyzer, which successfully removed the high levels of ammonia nitrogen background interference from physiological samples, plus a potentiometric ammonia gas detector [22]. This electrode-based, ammonia-liberating ASNase assay system enabled the quick and accurate concentration analysis of L-asparagine in blood serum samples up to 10−4 M levels [22]. In 1995, an ASNase-based biosensor was designed for ammonia detection via the combination of ASNase in garlic tissue cells, responsible for L-asparagine transformation into ammonia, subsequently detected by an ammonia gas electrode. This biosensor displayed a L-asparagine detection limit between 10−4 and 10−1 M [23]. Throughout the years, ASNase-based biosensors have been developed towards the detection of ammonia due to L-asparagine hydrolysis used to monitor L-asparagine in ALL patients, along with L-asparagine concentration analysis in foods (Table 1; Figure 2). Other biosensors were further developed, as detailed below.
Table 1. Industry, application, detection methodology, ASNase source and immobilization supports of ASNase-based biosensors.
|
Industry |
Application |
Detection Methodology |
ASNase1 Source |
Immobilization Supports |
Ref. |
|
__ |
Ammonia sensing |
Potentiometric ammonia gas detector |
Escherichia coli |
― |
[22] |
|
__ |
Ammonia sensing |
Ammonia gas electrode |
Garlic tissue cells |
― |
[23] |
|
__ |
Ammonia sensing |
Polypyrrole probe |
E. coli |
― |
[24] |
|
Pharmaceutical |
L-asparagine |
Phenol red |
Nitrocellulose membrane; silicone gel; calcium alginate beads |
[25] |
|
|
Food |
L-asparagine |
Phenol red |
Coliform |
Tetramethyl orthosilicate sol-gel |
[26] |
|
Ammonia sensing |
Ammonium-selective glass electrode |
Archaeoglobus fulgidus |
― |
[27] |
|
|
Food; pharmaceutical |
L-asparagine |
Nessler’s reagent |
Erwinia carotovora |
Plastic cuvette |
[28] |
|
96-well microplate |
[29] |
||||
|
Pharmaceutical |
L-asparagine |
Phenol red |
Capsicum |
Gelatin; polyacrylamide; agar; calcium alginate beads |
[30] |
|
|
Citrus limon |
Agar; agarose; gelatin; polyacrylamide; calcium alginate beads |
[31] |
||
|
|
Withania |
Gelatin; agarose; agar; calcium alginate beads |
[21] |
||
|
|
Cannabis sativa |
Gelatin; agarose; agar; calcium alginate beads; Whatman filter paper; hydrosol gel on nylon membrane |
[32] |
||
|
|
Catharanthus roseus |
Agar; soil; clay; k-carrageenan |
[11] |
1ASNase—L-asparaginase; 2ALL—acute lymphoblastic leukemia.
Figure 2. ASNase-based biosensors: ASNase sources, detection methodologies and immobilization supports.
2.1. ASNase from Bacteria for the Development of Biosensors
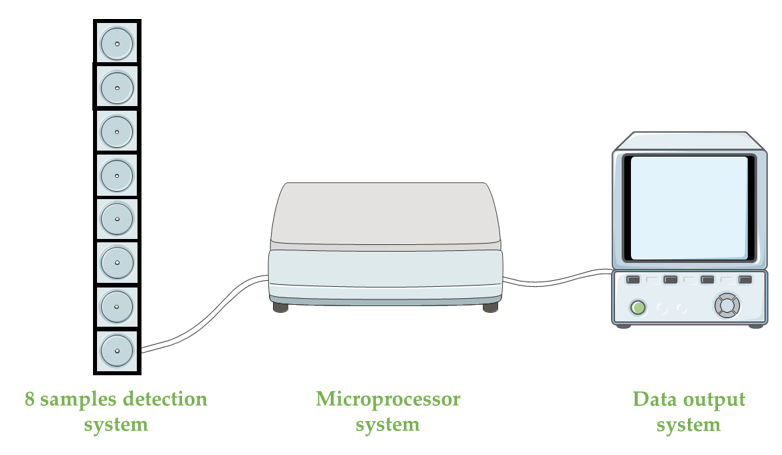
Even though ASNase is broadly distributed in nature (plants, animals, tissues and microorganisms (bacteria, filamentous fungi and yeast)), the majority of commercialized ASNase are from recombinant microorganisms, i.e.,Escherichia coli and Erwinia chrysanthemi [6,33,34]. Thus, a simple, safe and real time method suitable for biological applications, based on E. coli ASNase, was produced for ammonia detection by the connection of a conducting polymer, polypyrrole (PPY) probe, previously chemically deposited on the polyimide surface to a piezoelectric quartz crystal (PQC) in multi-channel series (MS) (PPY-MSPQC system) [24]. This method comprises three main elements: an eight-sample detection system, a microprocessor system (responsible for the signal transference from the frequency counter to a computer) and the data output system (Figure 3). In fact, the PPY-MSPQC system enabled an ASNase activity assay with the same accuracy as Nessler’s reagent method due to its excellent ammonia sensing features [24].
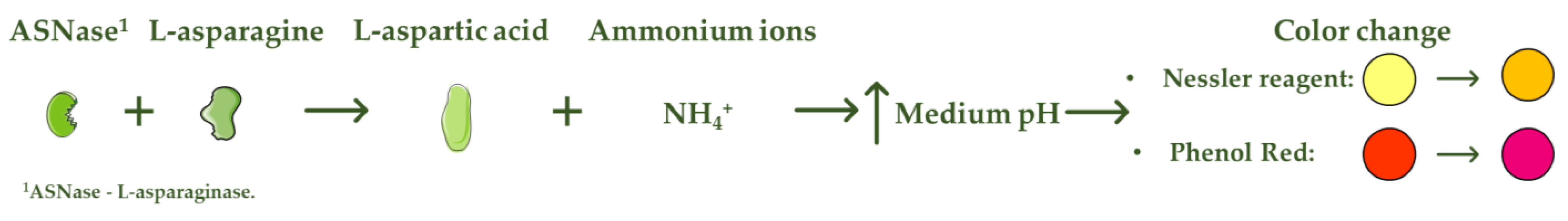 Figure 13.
Figure 13. Schematic diagram of the polypyrrole probe, deposited on the polyimide surface to a piezoelectric quartz crystal in multi-channel series (PPY-MSPQC system).
2.1.1. Pharmaceutical Industry
An E. coli ASNase-based colorimetric biosensor was established for L-asparagine monitoring in ALL patients via nitrocellulose membrane, silicon gel and calcium alginate beads methods (Table 2) [25]. These methods included the co-immobilization of ASNase solution (0.16 U) and pH indicator phenol red on nitrocellulose membranes, silicon gel coated on glass slides and sodium alginate solution (3%), respectively. While silicon gel showed a L-asparagine detection limit between 10−10 and 10−1 M, calcium alginate beads and nitrocellulose membrane displayed a smaller range of detection (10−9 – 10−1 M and 10−1 M, respectively). The production of ammonium ions (and L-aspartic acid) by L-asparagine deamination led to a color change from red to violet, which ranged from 5 seconds (s) to 7 minutes (min) and 10 s for 10−1 M; 1 to 2 min for 10−5 M; and 3 to 4 min for 10−10 M. In particular, calcium alginate beads presented color change response times of 2 min and 35 s for healthy blood serum samples (10−4 M) and 3 min and 10 s for leukemia blood serum samples (10−2 M), whose biocomponent remained stable for 25 to 30 days [25].
Table 2. Colorimetric ASNase-based biosensors: detection methodologies, medium conditions and color change.
|
Detection Methodologies |
Medium Conditions |
Color Change |
Ref. |
||
|
Phenol Red |
↑ pH |
|
|
|
[11,21,25,26,30–32] |
|
Nessler reagent |
↑ NH4+ |
|
|
|
[28,29] |
2.1.2. Food Industry
Although most ASNase-based biosensors have been applied to the pharmaceutical industry, certain developed biosensors have been used in the food industry. A fiber optic biosensor developed through the co-immobilization of ASNase producing Coliform bacterial cells, previously isolated from Fortis Hospital, Mohali (India) wastewater and phenol red onto circular plastic discs using tetramethyl orthosilicate (TMOS) sol-gel, was applied to study the L-asparagine concentration of four drinks, namely, tea, pineapple plus mango juice and wine [26]. The biosensor displayed a 7-min response time, detection limit of 1 × 10−9 M and a biocomponent storage stability of 40 days at 4 °C [26]. To surpass a major limitation of enzyme-based biosensors, i.e., the low stability of enzymes, a biosensor for L-asparagine was developed through the immobilization of a thermostable recombinant ASNase from Archaeoglobus fulgidus expressed in E. coli as a fusion protein in front of an ammonium-selective glass electrode, able to operate at high temperatures [27]. This biosensor with increased stability displayed a L-asparagine detection limit of 6 × 10−5 M, enabling its use in the food industry. In fact, ASNase remained stable at 85 °C, keeping 70% of its activity, in addition to showing a higher affinity for L-asparagine at 70 °C (Michaelis constant (KM) of 5 × 10−6) than at 37 °C (KM of 8 × 10−5). This was the first report of a potentiometric biosensor applying a thermostable ASNase [27]. Nevertheless, ASNase with negligible glutaminase activity is required since L-glutamine is an amino group donor for numerous biosynthetic reactions [28,35]. Prolonged low L-glutamine levels impair several biochemical functions, mainly in the liver [28,35]. While ASNase type I is a homodimeric cytosolic constitutive enzyme with high specific activity towards L-glutamine, plus relatively low affinity for L-asparagine, ASNase type II meets all the requirements since it is a homotetrameric periplasmic enzyme with high L-asparagine affinity, plus low activity towards L-glutamine, which is secreted only as a response to exposure to low nitrogen concentrations [6,34]. Instead, an ASNase cuvette-based biosensor was developed through the immobilization of a variant of ASNase from Erwinia carotovora with a single point mutation (Leu71Ile), which showed negligible glutaminase activity, via cross-linking with glutaraldehyde onto the inner surface of a plastic cuvette [28]. However, this biosensor displayed low ASNase activity in addition to low stability [28,29]. Thus, a highly specific ASNase microplate-based biosensor was designed by applying recombinant ASNase from E. carotovora expressed in E. coli, which showed high ASNase, along with low L-glutaminase activity [29]. ASNase extracted from E. carotovora and expressed in E. coli was initially mixed with bovine serum albumin (BSA), which functioned as the enzyme carrier, followed by cross-linking using glutaraldehyde, and deposition of the mixture into the wells of a 96-well microplate. While immobilized ASNase kept half of its initial activity within 30 days, free ASNase lost its activity following 8 days at 4 °C. Moreover, the biosensor presented a L-asparagine detection limit of 10 × 10−6 M, allowing around 20 cycles of sample analysis before total deterioration (ASNase activity below 60% of its initial value). This biosensor enabled micro-volume and high-throughput L-asparagine monitoring in foods, e.g., potatoes (720 ± 35 µg.g−1 to 1560 ± 45 µg.g−1 fresh weight), cheese, juices and asparagus [29].
2.2. ASNase from Plant Species for the Development of Biosensors
As depicted in Table 1, ASNase is present in certain plant species, such as Capsicum annum, Citrus limon, Withania somnifera, Cannabis sativa and Catharanthus roseus, enabling the development of various plant-ASNase-based colorimetric biosensors (Table 2) [11,21,23,30–32]. Table 3 summarizes the plant-ASNase-based biosensors, immobilization supports, L-asparagine detection limit, response time range, response time for leukemic blood serum samples and biocomponent stability found in the literature. The co-immobilization of ASNase extracted from the plant C. annum and phenol red via gelatin, polyacrylamide, agar and calcium alginate beads created a cost-effective diagnostic ASNase-based biosensor for L-asparagine monitoring in ALL patients [30]. The biocomponent was coupled with an ammonium ion-selective electrode responsible for the potential detection across its membrane. The immobilization methods include the addition of ASNase solution (0.5 U) to gelatin aqueous solution, acrylamide and bis-acrylamide solution (10%), agar solution (4%) or sodium alginate slurry (3%), followed by phenol red. While all methods displayed a L-asparagine detection limit between 10−9 and 10−1 M, their response time ranged from 10 to 21.6, 10 to 20, 7.5 to 14.2 and 7.1 to 12.3 s, whereas the biocomponent remained active for more than 15 days, 1 month, 15 days and 4 months for gelatin, polyacrylamide, agar, plus calcium alginate beads, respectively. Since the calcium alginate beads presented the fastest time response, in addition to stability for 4 months, this method was applied for L-asparagine detection in healthy (10−4 M) and leukemic blood serum samples (10−2 M), displaying color change response times of 9.4 and 11.2 s, respectively [30]. An alternative plant-ASNase-based biosensor for L-asparagine monitoring in ALL patients was developed through co-immobilization of ASNase extracted from the plant C. limon and phenol red via agar, agarose, gelatin, polyacrylamide and calcium alginate beads methods [31]. These methods consist of the addition of ASNase solution (0.3 U) to agar solution (4%), agarose solution (1.5%), gelatin aqueous solution (10%), acrylamide and bis-acrylamide solution (10%) or sodium alginate slurry (3%), followed by phenol red. All methods showed an L-asparagine detection limit between 10−10 and 10−1 M, while the color change response times were 11, 14, 16, 15, and 9.3 s for healthy (10−4 M), plus 13, 16, 20, 18, and 11 s for leukemia blood serum samples (10−2 M), respectively. The biocomponent remained stable for 1 month, 25, 9 and 25 days, plus 3 months in agar cakes, agarose pieces, gelatin, polyacrylamide gel blocks and calcium alginate beads, respectively. Among these, calcium alginate beads were selected as the best immobilization method [31]. Further developments in cost-effective plant-ASNase-based biosensors for L-asparagine monitoring in ALL patients was achieved by the co-immobilization of ASNase extracted from the plant W. somnifera and phenol red through gelatin, agarose, agar and calcium alginate beads methods [21]. Immobilization methods comprised the addition of ASNase solution (0.5 U) to gelatin aqueous solution (10%), agarose solution (1.5%), agar solution (4%) or sodium alginate slurry (3%), followed by phenol red. All methods presented a L-asparagine detection limit between 10−10 and 10−1 M. Color change response times were 16, 13, 11 and 9.3 s for healthy blood serum samples (10−4 M) and 19, 15, 12 and 11 s for leukemic blood serum samples (10−2 M), respectively. Since the biocomponent remained stable for more than 4, 12, 4 days and 2 months, respectively, along with the lowest response time, calcium alginate was selected as the best method for L-asparagine detection [21].
A different cost-effective plant-ASNase-based biosensor was created by the co-immobilization of ASNase extracted from C. sativa and phenol red using the same immobilization methods, along with Whatman filter paper and hydrosol gel on nylon membrane methods [32]. While all methods displayed the same L-asparagine detection limit (10−10–10−1 M), the response time of the biosensors ranged from 9 to 21 s for leukemic blood serum samples (10−2 M) and from 7.25 to 16 s for healthy blood serum samples (10−5 M). Hydrosol gel on nylon membrane was selected as the best method due to its fastest response time for both leukemic and healthy blood serum samples (9 and 7.25 s). Furthermore, ASNase remained active in air tight containers for more than 4 months after drying [32]. Additionally, ASNase extracted from C. roseus was co-immobilized with phenol red via agar, soil, clay and k-carrageenan methods [11]. Although all methods showed a L-asparagine detection limit ranging from 10−10 to 10−1 M, their response time varied from 7 to 14, 4 to 12, 3 to 11 and 3 to 10 s for agar, soil, clay and k-carrageenan, respectively. Therefore, the k-carrageenan method was selected for the development of a plant ASNase-based biosensor, which enabled L-asparagine concentration quantification in leukemic (10−3–10−2 M) and healthy blood serum samples (10−6–10−5 M) [11]. However, to date, we believe C. sativa-based biosensor using the hydrosol gel on nylon membrane method is the best plant-ASNase-based biosensor since it presented the second lowest response time for leukemia blood serum samples (9 s) in addition to having the highest biocomponent stability (more than 4 months).
All ASNase-based biosensors reported the use of blood serum samples, showing that the interference of the matrix has been considered and can be overcome. However, further studies are required to validate the use of whole blood samples able to overcome interferences of this more complex matrix. On the other hand, biofouling should be considered as well, i.e., the spontaneous macromolecules or microorganisms accumulation, which can physically limit the target analyte diffusion to the sensor surface [36–38]. If that is the case, strategies such as antifouling surface coatings with high biocompatibility, which inhibit biofouling through hydrophilic interactions, steric or electrostatic repulsions, plus low surface energy or physical (e.g., physical adsorption, mechanical coating, superhydrophobic, nanoporous structure) plus chemical (e.g., self-assembled monolayers and polymer brushes) strategies must be developed [39].
Table 3.
Plant-ASNase-based biosensors, immobilization supports, L-asparagine detection limit (M), response time range (s), response time for leukemic blood serum samples (s) and biocomponent stability.
|
Plant-ASNase1-Based Biosensors |
Immobilization Supports |
L-Asparagine Detection Limit (M) |
Response Time Range (s) |
Response Time for Leukemic Blood Serum Samples (s) |
Biocomponent Stability |
Ref |
|
C. annum-based biosensor |
Gelatin |
10−9–10−1 |
10–21.6 |
20 |
>15 days |
[30] |
|
Polyacrylamide |
10–20 |
18.7 |
>1 month |
|||
|
Agar |
7.5–14.2 |
12.5 |
>15 days |
|||
|
Calcium alginate beads |
7.1–12.3 |
11.2 |
>4 months |
|||
|
C. limon-based biosensor |
Agar |
10−10–10−1 |
6–14.2 |
13 |
1 month |
[31] |
|
Agarose |
9–16.4 |
16 |
25 days |
|||
|
Gelatin |
10–22 |
20 |
9 days |
|||
|
Polyacrylamide |
10–20 |
18 |
25 days |
|||
|
Calcium alginate beads |
7–12 |
11 |
3 months |
|||
|
W. somnifera-based biosensor |
Gelatin |
10−10–10−1 |
10–22 |
19 ± 0.5 |
>4 days |
[21] |
|
Agarose |
10–17 |
15 |
>12 days |
|||
|
Agar |
7–14 |
12 |
>4 days |
|||
|
Calcium alginate beads |
7–12 |
11 |
>2 months |
|||
|
C. sativa-based biosensor |
Gelatin |
10−10–10−1 |
8–21 |
19 |
― |
[32] |
|
Agarose |
9.17–16 |
15.8 |
― |
|||
|
Agar |
7.3–15 |
13.3 |
― |
|||
|
Calcium alginate beads |
7–11 |
11.1 |
― |
|||
|
Whatman filter paper |
11–23 |
21 |
― |
|||
|
Hydrosol gel on nylon membrane |
5–10 |
9 |
>4 months |
|||
|
C. roseus-based biosensor |
Agar |
10−10–10−1 |
7–14 |
― |
― |
[11] |
|
Soil |
4–12 |
― |
― |
|||
|
Clay |
3–11 |
― |
― |
|||
|
k-carrageenan |
3–10 |
7 |
― |
1ASNase—L-asparaginase.
- Conclusions and Future Perspectives
ASNase-based biosensors have been increasingly emerging, not only for pharmaceutical applications through the L-asparagine monitoring in blood serum samples, but in the food industry via L-asparagine concentration analysis in foods as well. These biosensing systems display high potential, which is due to their low-cost, simplicity, ease of use, micro-volume, nanolevel L-asparagine detection, fast response and high biocomponent stability.
Thus far, the best developed biosensor within the pharmaceutical field is the C. sativa-based biosensor applying the hydrosol gel on nylon membrane method due to displaying the second lowest response time for leukemia blood serum samples (9 s), in addition to having the highest biocomponent stability (more than 4 months). On the other hand, regarding the food industry, the highly specific ASNase microplate-based biosensor with recombinant ASNase from E. carotovora expressed in E. coli should be highlighted, which kept half of its initial activity within 30 days at 4 °C, allowing around 20 cycles of sample analysis before total deterioration, enabling micro-volume and high-throughput L-asparagine monitoring in potatoes, cheese, juices and asparagus.
Based on these promising results, it is expected the development of additional ASNase-based biosensors using alternative ASNase sources, immobilization supports and methods, leading to the design and subsequent low-cost and scalable production of cutting-edge biosensors. These should be designed to avoid interferences and be able to deal with more complex matrices, such as whole blood samples and more complex food samples. On the other hand, low-cost and novel devices whose mode of action is beyond the color change, as well as with higher accuracy and sensitivity for L-asparagine, along with higher operational stability, should be investigated as alternative strategies.
Author Contributions: J.C.F.N. and R.O.C., writing—original draft preparation; J.C.F.N., M.C.N., A.P.M.T. and M.G.F., conceptualization; V.C.S.-E., J.L.F., C.G.S., M.C.N., M.G.F. and A.P.M.T., review—editing and supervision. All authors have read and agreed to the published version of the manuscript.
Funding: This work was developed within the scope of the project CICECO—Aveiro Institute of Materials, UIDB/50011/2020 & UIDP/50011/2020, financed by national funds through the Portuguese Foundation for Science and Technology/MCTES. This work was also financially supported by Base Funding—UIDB/EQU/50020/2020 of the Associate Laboratory LSRE-LCM-funded by national funds through FCT/MCTES (PIDDAC), and POCI-01-0145-FEDER-031268-funded by FEDER, through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI), and by national funds (OE), through FCT/MCTES, and by FAPESP (2018/06908-8).
Acknowledgments: Ana P. M. Tavares acknowledges the FCT for the research contract CEECIND/2020/01867. Valéria C. Santos-Ebinuma acknowledges FAPESP for financial support. Raquel O. Cristóvão acknowledges FCT funding under DL57/2016 Transitory Norm Programme. João C. F. Nunes acknowledges SPQ and FCT for the PhD fellowship (SFRH/BD/150671/2020).
Conflicts of Interest: The authors declare no conflict of interest.
Entry Link on the Encyclopedia Platform:
References
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification 1 International Union of Pure and Applied Chemistry: Physical Chemistry Division, Commission I.7 (Biophysical Chemistry); Analytical Chemistry Division, Commission V.5 (Electroanalytical). Bioelectron. 2001, 16, 121–131, doi:10.1016/S0956-5663(01)00115-4.
- Bahadır, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Biochem. 2015, 478, 107–120, doi:10.1016/j.ab.2015.03.011.
- Turner, A.; Karube, I.; Wilson, G.S. Biosensors: Fundamentals and Applications, 1st ed.; Oxford University Press: Oxford, UK, 1987; ISBN 0198547242.
- Songa, E.A.; Okonkwo, J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: A review. Talanta 2016, 155, 289–304, doi:10.1016/j.talanta.2016.04.046.
- Zhilei, W.; Zaijun, L.; Xiulan, S.; Yinjun, F.; Junkang, L. Synergistic contributions of fullerene, ferrocene, chitosan and ionic liquid towards improved performance for a glucose sensor. Bioelectron. 2010, 25, 1434–1438, doi:10.1016/j.bios.2009.10.045.
- Nunes, J.C.F.; Cristóvão, R.O.; Freire, M.G.; Santos-Ebinuma, V.C.; Faria, J.L.; Silva, C.G.; Tavares, A.P.M. Recent strategies and applications for L-asparaginase confinement. Molecules 2020, 25, 5827, doi:10.3390/molecules25245827.
- Brena, B.; González-Pombo, P.; Batista-Viera, F. Immobilization of enzymes: A literature survey. In Immobilization of Enzymes and Cells; 2013; Volume 1051, pp. 15–31, ISBN 9781627035491.
- Kidd, J.G. Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum. I. Course of transplanted cancers of various kinds in mice and rats given guinea pig serum, horse serum, or rabbit serum. Exp. Med. 1953, 98, 565–582, doi:10.1084/jem.98.6.565.
- Kidd, J.G. Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum. II. Studies on the nature of the active serum constituent: Histological mechanism of the regression: Tests for effects of guinea pig serum on lymphoma cells in vitro. Exp. Med. 1953, 98, 583–606, doi:10.1084/jem.98.6.583.
- Ando, M.; Sugimoto, K.; Kitoh, T.; Sasaki, M.; Mukai, K.; Ando, J.; Egashira, M.; Schuster, S.M.; Oshimi, K. Selective apoptosis of natural killer-cell tumours by L-asparaginase. J. Haematol. 2005, 130, 860–868, doi:10.1111/j.1365-2141.2005.05694.x.
- Sandeep, P.; Raman, K.; Kuldeep, K. Enzyme based asparagine biosensor for the detection of asparagine levels in leukemic samples. J. Appl. Biol. Pharm. Technol. 2015, 6, 40–43.
- Avramis, V.I.; Panosyan, E.H. Pharmacokinetic/pharmacodynamic relationships of asparaginase formulations: The past, the present and recommendations for the future. Pharmacokinet. 2005, 44, 367–393, doi:10.2165/00003088-200544040-00003.
- Paillassa, J.; Leguay, T.; Thomas, X.; Huguet, F.; Audrain, M.; Lheritier, V.; Vianey-Saban, C.; Acquaviva-Bourdain, C.; Pagan, C.; Dombret, H.; et al. Monitoring of asparagine depletion and anti-L-asparaginase antibodies in adult acute lymphoblastic leukemia treated in the pediatric-inspired GRAALL-2005 trial. Blood Cancer J. 2018, 8, 45, doi:10.1038/s41408-018-0084-5.
- Cancer, I.A. for R. on; IARC monographs on the evaluation of carcinogenic risks to humans: WHO, Vol. 63. Dry cleaning, some chlorinated solvents and other industrial chemicals. Chim. Acta 1996, 336, 229–230, doi:10.1016/S0003-2670(97)89591-8.
- Joint Food and Agriculture Organization; World Health Organization. Health Implications of Acrylamide in Food: Report of a Joint FAO/WHO Consultation, WHO Headquarters, Geneva, Switzerland, 25–27 June 2002; World Health Organization: Geneva, Switzerland, 2002; ISBN 9241562188.
- Abt, E.; Robin, L.P.; McGrath, S.; Srinivasan, J.; DiNovi, M.; Adachi, Y.; Chirtel, S. Acrylamide levels and dietary exposure from foods in the United States, an update based on 2011–2015 data. Food Addit. Contam. Part. A Chem. Anal. Control. Expo. Risk Assess. 2019, 36, 1475–1490, doi:10.1080/19440049.2019.1637548.
- Tardiff, R.G.; Gargas, M.L.; Kirman, C.R.; Leigh Carson, M.; Sweeney, L.M. Estimation of safe dietary intake levels of acrylamide for humans. Food Chem. Toxicol. 2010, 48, 658–667, doi:10.1016/j.fct.2009.11.048.
- Xu, F.; Oruna-Concha, M.-J.; Elmore, J.S. The use of asparaginase to reduce acrylamide levels in cooked food. Food Chem. 2016, 210, 163–171, doi:10.1016/j.foodchem.2016.04.105.
- Parker, J.K.; Balagiannis, D.P.; Higley, J.; Smith, G.; Wedzicha, B.L.; Mottram, D.S. Kinetic model for the formation of acrylamide during the finish-frying of commercial french fries. Agric. Food Chem. 2012, 60, 9321–9331, doi:10.1021/jf302415n.
- Zubavichus, Y.; Fuchs, O.; Weinhardt, L.; Heske, C.; Umbach, E.; Denlinger, J.D.; Grunze, M. Soft X-Ray-induced decomposition of amino acids: An XPS, mass spectrometry, and NEXAFS study. Res. 2004, 161, 346–358.
- Kumar, K.; Kataria, M.; Verma, N. Plant asparaginase-based asparagine biosensor for leukemia. Cells Nanomed. Biotechnol. 2013, 41, 184–188, doi:10.3109/10731199.2012.716062.
- Fraticelli, Y.M.; Meyerhoff, M.E. On-Line gas dialyzer for automated enzymatic analysis with potentiometric ammonia detection. Chem. 1983, 55, 359–364, doi:10.1021/ac00253a039.
- Kim, S.J.; Kim, G.M.; Bae, Y.J.; Lee, E.Y.; Hur, M.H.; Ahn, M.K. Determination of L-asparagine using a garlic tissue electrode. Yakhak Hoeji 1995, 39, 113–117.
- Ren, J.; He, F.; Zhang, L. The construction and application of a new PPY-MSPQC for L-asparaginase activity assay. Actuators B Chem. 2010, 145, 272–277, doi:10.1016/j.snb.2009.12.006.
- Verma, N.; Kumar, K.; Kaur, G.; Anand, S. coli K-12 asparaginase-based asparagine biosensor for leukemia. Artif. Cells, Blood Substit. Biotechnol. 2007, 35, 449–456, doi:10.1080/10731190701460358.
- Verma, N.; Bansal, M.; Kumar, S. Whole cell based miniaturized fiber optic biosensor to monitor L-asparagine. Appl. Sci. Res. 2012, 3, 809–814.
- Li, J.; Wang, J.; Bachas, L.G. Biosensor for asparagine using a thermostable recombinant asparaginase from Archaeoglobus fulgidus. Chem. 2002, 74, 3336–3341, doi:10.1021/ac015653s.
- Kotzia, G.A.; Labrou, N.E. Engineering substrate specificity of carotovora L-asparaginase for the development of biosensor. J. Mol. Catal. B Enzym. 2011, 72, 95–101, doi:10.1016/j.molcatb.2011.05.003.
- Labrou, N.E.; Muharram, M.M. Biochemical characterization and immobilization of Erwinia carotovora L-asparaginase in a microplate for high-throughput biosensing of L-asparagine. Microb. Technol. 2016, 92, 86–93, doi:10.1016/j.enzmictec.2016.06.013.
- Kumar, K.; Walia, S. L-asparaginase extracted from Capsicum annum L and development of asparagine biosensor for leukemia. Transducers 2012, 144, 192–200.
- Kumar, K.; Punia, S.; Kaur, J.; Pathak, T. Development of plant asparagine biosensor for detection of leukemia. Pharm. Biomed. Sci. 2013, 35, 1796–1801.
- Teena, P.; Raman, K.; Jagjit, K.; Kuldeep, K. Isolation of L-asparaginase from Cannabis Sativa and development of biosensor for detection of asparagine in leukemic serum samples. J. Pharm. Technol. 2014, 7, 850–855.
- Cachumba, J.J.M.; Antunes, F.A.F.; Peres, G.F.D.; Brumano, L.P.; Dos Santos, J.C.; Da Silva, S.S. Current applications and different approaches for microbial L-asparaginase production. J. Microbiol. 2016, 47, 77–85, doi:10.1016/j.bjm.2016.10.004.
- Castro, D.; Marques, A.S.C.; Almeida, M.R.; de Paiva, G.B.; Bento, H.B.S.; Pedrolli, D.B.; Freire, M.G.; Tavares, A.P.M.; Santos-Ebinuma, V.C. L-asparaginase production review: Bioprocess design and biochemical characteristics. Microbiol. Biotechnol. 2021, 105, 4515–4534, doi:10.1007/s00253-021-11359-y.
- Sokolov, N.N.; Eldarov, M.A.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Abakumova, O.Y.; Podobed, O.V.; Melik-Nubarov, N.S.; Kudryashova, E.V.; Grishin, D.V.; Archakov, A.I. Bacterial recombinant L-asparaginases: Properties, structure, and anti-proliferative activity. Moscow Suppl. Ser. B 2015, 9, 325–338, doi:10.1134/S199075081504006X.
- Xu, J.; Lee, H. Anti-biofouling strategies for long-term continuous use of implantable biosensors. Chemosensors 2020, 8, 66, doi:10.3390/chemosensors8030066.
- Wisniewski, N.; Moussy, F.; Reichert, W.M. Characterization of implantable biosensor membrane biofouling. Fresenius J. Anal. Chem. 2000, 366, 611–621, doi:10.1007/s002160051556.
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme biosensors for biomedical applications: Strategies for safeguarding analytical performances in biological fluids. Sensors 2016, 16, 780,, doi:10.3390/s16060780.
- Lin, P.-H.; Li, B.-R. Antifouling strategies in advanced electrochemical sensors and biosensors. Analyst 2020, 145, 1110–1120, doi:10.1039/c9an02017a.