A number of insights derive from viewing flipons as scaffolds for condensates. Flipons provide a controlled way to initiate condensate formation, one subject to natural selection. The alternative conformation localizes required factors needed to regulate transcription, RNA processing, and epigenetic modification, while excluding nucleosomes and other B-DNA- and A-RNA-specific proteins that produce competing outcomes.
- Z-DNA
- Z-RNA
- flipons
- simple repeats
- condensates
- G4
- evolution
- MYC
- non-coding RNA
- DNA conformation
- complexity
- phase separation
- enhancersome
- nucleosome
1. Starting Simple
The theme of this article is presented in Figure 1 . Here, a nucleic acid structural motif is recognized by a structure-specific protein interaction. The nucleic acid acts as a scaffold and the protein as an anchor for cellular machines. Patches on the anchor protein provide a docking site for other proteins ( Figure 1 , left panel). The outcome depends on the functions of the assembled proteins, any one of which may have peptide patches for the attachment of additional proteins. In the simplest case, both the nucleic acid structures and the peptide patches are encoded by simple sequence repeats (SSRs) ( Figure 1 , right panel). The approach to build complex biological machines is adaptative for ever-changing environments.
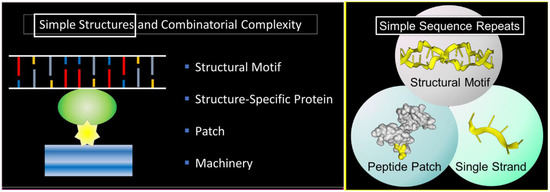
The scheme exploits the properties of SSRs that enable them to encode alternative nucleic acid structures, called flipons, to specify simple peptide patches that fold in different ways and to engage sequence-specific, single-stranded binding proteins that play multiple roles in condensate biology. We will focus in this review on the biology of flipons and peptide patches: how they can initiate and how they can promote condensate formation to optimize responses. We will discuss the role of SSRs in disease and their evolutionary impact. To introduce the concepts, we will start with a description of condensates and their role in classical genetics and then move on to SSRs and flipon genetics.
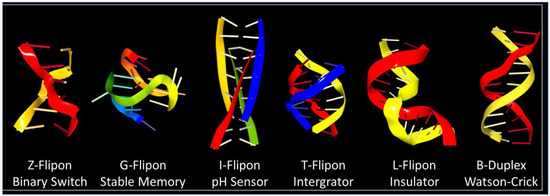
2. Z-Flipons
Z-RNA regulates both type I interferon responses by adenosine deaminase RNA specific (ADAR1, encoded by ADAR) [1][22] and the programmed cell death necroptosis pathway by Z-DNA binding protein 1 (ZBP1) [2][44]. In both cases, the left-handed helix is recognized by a structure specific, winged helix-turn-helix Zα protein domain without any base-specific contacts involved [3][4][45,46]. With ADAR1, Zα and another structure-specific, double-stranded RNA (dsRNA)-specific binding motif target the deaminase domain to modify adenosines in dsRNA, forming inosine that is treated by the downstream processes as the equivalent of guanosine. This process produces non-synonymous codon changes in a limited number of human substrates [5][47].
It is likely that other Z-binding proteins exists, some of which may also bind B-DNA. Simple peptide repeats, such as peptides with alternating lysine residues, have high specificity for Z-DNA when tested with a methylated polymer [6][48]. These repeats that are likely IDRs are present in a number of interesting proteins. One example is the DNA methyl transferase I (encoded by DNMT1) that contains a lysine–glycine repeat, 10 amino acids long. The repeat is unstructured in the native DNMT1 crystal (Protein Data Bank (PDB): 5GUV) but interacts with ubiquitin-specific peptidase 7 (USP7) in the co-crystal (PDB: 4YOC). USP7 negatively modulates the activity of DNMT1 [7][49]. It is also possible that the repeat serves to localize DNMT1 to regions that form Z-DNA to methylate them. Another example is the histone H2A.Z variant 2 (encoded by H2AZ2; NCBI: NP_036544.1) that contains three alternating lysine–alanine repeats and destabilizes nucleosomes.
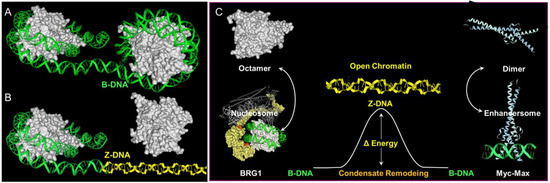
Less clear cut are the roles of poly-lysine repeats in the large ATP-dependent SWI/SNF chromatin-remodeling proteins BRG1 (encoded by SMARCA4) and BRM (encoded by SMARCA2) ( Figure 23 ). The SWI/SNF complex is able to eject nucleosomes from DNA. The stress of DNA negative supercoiling, previously relieved by winding the DNA around the histone octamer, now becomes available to stabilize Z-DNA ( Figure 23 ). Here, the left-handed solenoidal winding around the nucleosome is converted into a left-handed DNA twist. Previous reports have revealed that Z-DNA formation induced by BRG1 is associated with activation of the colony-stimulating factor 1 (CSF1) gene [8][50] and also the transcription of the heme oxygenase 1 gene (encoded by HMOX1) induced by nuclear factor, erythroid 2 like 2 (NRF2, encoded by NFE2L2) in response to oxidative stress [9][51]. In the CSF1 promoter, Z-DNA formation occurs while DNA is bound by the nucleosome [10][52], consistent with a model where local induction of Z-formation in a Z-prone sequence by BRG1 initiates nucleosome eviction to create a region of open chromatin. As the flip from B-DNA to Z-DNA is cooperative, the propagation of Z-formation into adjacent segments will dislodge the entire octamer, releasing negative supercoiling that further promotes Z-formation in the region ( Figure 23 ) and the binding of structure-specific proteins to that locus. The energy stored as Z-DNA is then available to promote the assembly of new protein complexes on the DNA. The flip to Z-DNA controls both the location and the timing of subsequent events, producing a switch from one genetic program to another. The flip back to B-DNA relieves topological stresses as the new condensate forms.
3. G-Flipons
G-flipons can fold in a number of different ways, with strands running either parallel or anti-parallel with loops, providing connections that vary in length and position. The G4 quartet is recognized in a number of different ways [11][53]. Crystal structures of DDX36 and RAP1 proteins bound to G4 DNA reveal that the G4 caps are contacted by hydrophobic residues present in an α-helix, with engagement of the phosphate backbone by basic residues [12][13][54,55]. RAP1 also binds B-DNA through the same helix that it uses to bind G4, but through a different face [13][55]. A number of other proteins initially characterized as single-stranded binders have subsequently been shown to bind G4 with high affinity. While their single-strand specificity was evident because they contain the RNA recognition motif (RRM), the role of peptide patches that recognizes G4 was not appreciated initially fell within IDR, lacking structure [14][56]. In these cases, the entropic cost of binding a disordered, single-stranded RNA is reduced by the RRM structure, while the entopic cost of docking to a peptide patch is lessened by the prepositioned backbone of the G4 motif [15][57]. Recognition of G-flipons through arginine– glycine motifs within IDR is common [16][37], with hydrophobic residues such as tyrosine and phenylalanine [14][17][56,58] showing different preferences for G4 RNA and G4 DNA [18][59]. Other modes of binding to G4 structures also exist. Fragile mental retardation protein recognizes the junction between B-DNA and the G4 structure, combining backbone contacts with base-specific ones [19][60].
The potential biological roles for G-flipons are various. Some are evidenced by both genetic and biochemical studies. There are mendelian diseases caused by defects in DNA repair and replication. The helicase variants involved show altered binding to G4 in vitro [20][61]. The 425 G4 interacting proteins recently identified using probes containing various constrained G4 structures are enriched for spliceosomes, RNA transport, RNA degradation, mRNA surveillance, DNA replication, and homologous recombination pathways. One G4-binding protein complex, the negative elongation factor (NELF), regulates gene expression in eukaryotic cells by promoting RNA polymerase pausing [21][62]. Other cell-based studies demonstrate the presence of nuclear G4 structures [22][23][63,64]. Collectively, the above studies suggest an important role for G-flipons in localizing cellular machines to regions where outcomes modify both normal function and disease risk [24][65].
4. When Flipons and Codons Clash
Many simple repeats that undergo expansion produce autosomal dominant disease [25][81], providing insight into their biology. The adverse outcomes reflect the abilities of repeats to both form alternative structures and encode longer peptide patches that seed aggregates. For example, the hexameric FTD/ALS repeat sequence GGGGCC can be transcribed from both strands to form a G4 quartet from one [26][82] and an I-motif ( Figure 32 ) from the other [27][83]. The RNA transcribed from both strands is translated without the need for a traditional AUG start codon, a process called repeat-associated non-AUG (RAN) translation [28][84]. The RNA produced encodes six different dipeptide protein products, depending on the reading frame, with the two containing arginine being the most toxic. Disruption of many fundamental processes by the alternative nucleic conformations, by the peptide repeats, and by loss of function of the protein have been proposed as causes of disease [29][85]. Here, the alternative flipon conformations locked in by these diseases nucleate condensates that are only formed transiently within normal cells. The condensates persist. The proteins involved may be critical to switching one cellular response to another or may be involved in a separate pathway that is disrupted by their sequestration. The outcomes vary with the functions of the flipons involved.
The question arises whether flipon sequences also encode peptide repeats that bind to the flipon that encodes them. The CGG repeat expansion that causes fragile X-related tremor/ataxia syndrome (FXTAS) does generate a polyglycine peptide (PPG) that appears to stabilize a G4 quartet formed by the RNA transcribed [32][86], with a PPG longer than nine residues by itself being insoluble [33][87]. In this case, the simple repeat codes for an alternative RNA conformation and a single amino acid repeat peptide that also forms higher-order protein structures. The interaction of the peptide with the RNA leads to disease. In cells, both PPG and G4 partition together into granules that stain with a G4-specific antibody. The granules also accumulate many other proteins, potentially interfering with the formation of alternative complexes that are essential for normal cell function. Here, codons and flipons clash. The two different schema for the encoding of genetic information, one that specifies the amino acid sequence and the other that influences nuclear acid conformation, directly target each other, producing a negative outcome. RNA from a simple repeat expansion causes disease in FXTAS by undergoing RAN translation to produce peptides that locks the flipon sequence encoding the peptide into an alternative conformation, preventing the production of more peptide, which leads to the accumulation of more RNA that then is translated into new peptide. The self-referential and self-sustaining nature of this system produces a futile cycle, leading to system failure.
