Canine adenoviruses (CAdVs) can be the backbones of viral vectors that could be applied in recombinant vaccines or for gene transfer in dogs and in serologically naïve humans. Although conventional plasmid-based reverse genetics systems can be used to construct CAdV vectors, their large genome size creates technical difficulties in gene cloning and manipulation. Here, we established an improved reverse genetics system for CAdVs using bacterial artificial chromosomes (BACs), in which genetic modifications can be efficiently and simply made through BAC recombineering. Our established BAC-based reverse genetics system for CAdVs would be a useful and powerful tool for basic and advanced practical studies with these viruses.
- canine
- adenovirus
- reverse genetics
- vector
- bacterial artificial chromosome
1. Definition
Canine adenoviruses (CAdVs) are divided into pathotypes CAdV1 and CAdV2, which cause infectious hepatitis and laryngotracheitis in canid animals, respectively. They can be the backbones of viral vectors that could be applied in recombinant vaccines or for gene transfer in dogs and in serologically naïve humans. Although conventional plasmid-based reverse genetics systems can be used to construct CAdV vectors, their large genome size creates technical difficulties in gene cloning and manipulation.
2. Introduction
Canine adenovirus (CAdV), a double-stranded DNA virus that belongs to the genus Mastadenovirus in the family Adenoviridae, is divided into pathotypes CAdV1 and CAdV2, the former of which causes infectious hepatitis and encephalitis in dogs, bears, red foxes, and wolves [1][2][3], and the latter causes infectious laryngotracheitis in dogs [4]. The molecular basis of this pathogenic difference remains unknown. Outbreaks of CAdV infections in wildlife and animal shelters have been reported in recent years [5][6]. To protect domestic dogs from infections by this virus, attenuated CAdV2 vaccines, which are also effective against serologically cross-reactive CAdV1 infections, are widely used.
Currently, several adenovirus vectors are being commonly employed and tested for clinical use. The replication-defective constructs, in which essential genes (e.g., early region 1 (E1)) are replaced with foreign therapeutic or protective antigen genes, can be used for gene and cancer therapies and as recombinant vaccines, whereas the replication-competent constructs, which preferentially replicate in cancer cells only, can be applied for oncolytic therapy [7][8]. Several human adenovirus (HAdV)-based vectors for gene transfer have been evaluated for clinical use in humans for many years. However, any pre-existing cellular or humoral immunity against HAdV can prevent efficient gene transfer and affect the duration of gene expression. By contrast, humans do not have any pre-existing immunity against CAdVs [9], which presents CAdV-based vectors as promising for human use. Additionally, CAdV can potentially be an oncolytic virus for canine cancers [10].
The reverse genetics system, through which an intended gene manipulation can be introduced into the viral genome, is a powerful tool for use in basic studies on CAdVs (e.g., on their pathogenesis) as well as in practical studies (e.g., on CAdV-based vectors and recombinant vaccines). Several techniques to construct recombinant AdVs, including CAdVs, have been reported. In vitro ligation or its improved method, in which foreign DNA is directly ligated to AdV DNA digested with restriction enzyme, requires a unique restriction enzyme site and has low ligation efficiency [11][12][13][14]. The mammalian cell-based homologous recombination method, in which foreign DNA flanked by homology arms and adenovirus DNA are introduced into cells, requires long homology arms and repeated plaque purifications [15][16]. The bacteria-based homologous recombination method, in which the full-length AdV genome is cloned into plasmid via homologous recombination in the Escherichia coli BJ5183 strain, is widely used to construct infectious clones and to manipulate viral DNA. However, this method also requires long homology arms and a unique restriction enzyme site [9][17][18][19][20][21]. The yeast artificial chromosome-based method, which requires an additional yeast host, is infrequently used [22].
3. Construction of Bacterial Artificial Chromosome Clones of CAdV1 and CAdV2
To construct infectious BAC clones of CAdVs, we first determined the full-length sequences of the CAdV1 (strain D43) and CAdV2 (strain A2) genomes using next-generation sequencing, indicating 30,533 bp and 31,320 bp as the full lengths and mapping their identical gene arrangements to those of the reference viruses, respectively. Consequently, pBAC-CAdV1-ITR-GalK and pBAC-CAdV2-ITR-GalK were constructed, and the full-length viral genomes were then inserted into the BAC vector through homologous recombination with Red recombinase in SW102 cells. After the SW102 cell colonies had been screened by PCR, pBAC-CAdV1 and pBAC-CAdV2 were obtained (Figure 1A), and their proper constitutions were confirmed from their restriction enzyme digestion patterns (Figure 1B).
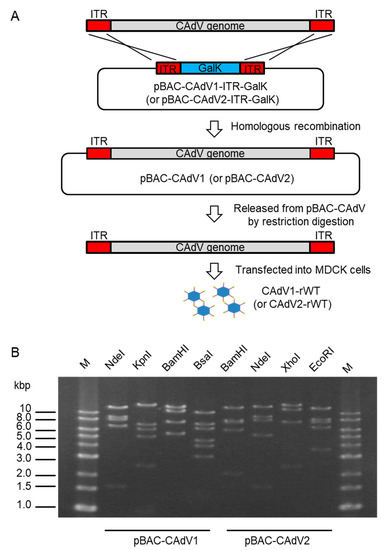
Figure 1. Cloning of canine adenovirus (CAdV) genomes into bacterial artificial chromosome (BAC) vectors. (A) CAdV1 (or CAdV2) DNA was introduced into SW102 cells harboring pBAC-CAdV1-ITR-GalK (or pBAC-CAdV2-ITR-GalK). The plasmid contains a galactokinase (galK) expression cassette flanked by approximately 50 bp of inverted terminal repeats (ITR) of the viral genome. After homologous recombination by Red recombinase, cells were selected using GalK, and pBAC-CAdV1 (or pBAC-CAdV2) was obtained. Then, this plasmid was digested with a restriction enzyme and transfected into Madin–Darby canine kidney (MDCK) cells, following which the recombinant virus CAdV1-rWT (or CAdV2-rWT) was rescued. (B) Restriction enzyme analysis of pBAC-CAdV1 and pBAC-CAdV2. pBAC-CAdV1 was digested with NdeI, KpnI, BamHI, and BsaI, whereas pBAC-CAdV2 was digested with BamHI, NdeI, XhoI, and EcoRI. The predicted molecular weights of the digested fragments of pBAC-CAdV1 are 13099, 8710, 8038, 6737, and 1603 bp for NdeI; 16319, 7077, 6210, 5110, 2490, and 981 bp for KpnI; 13573, 10982, 7620, 5549, and 463 bp for BamHI; and 10530, 7212, 6386, 4670, 4041, 3117, 825, 493, 471, 153, 147, and 142 bp for BsaI. The predicted molecular weights of the digested fragments of pBAC-CAdV2 are 13521, 7998, 7620, 6151, 2109, 851, and 722 bp for BamHI; 14820, 9059, 8127, 5363, and 1603 bp for NdeI; 16628, 12194, 7688, and 2462 bp for XhoI; and 13014, 8192, 7592, 6532, and 3642 bp for EcoRI. M: 1-kb DNA ladder marker.
4. Generation and Growth Properties of Recombinant CAdV1 and CAdV2
The CAdV DNAs were released from pBAC-CAdV1 or pBAC-CAdV2 by restriction enzyme digestion and transfected into MDCK cells. Then, infectious viruses (CAdV-rWTs) showing clear cytopathic effects were obtained from the transfected cells, following one additional passage with MDCK cells. The restriction enzyme digestion patterns of the CAdV-rWT genomes were identical to those of the CAdV-WTs (Figure 2A,B), suggesting a lack of major DNA rearrangement in the recombinant viruses. Additionally, the growth properties of the CAdV-rWTs were similar to those of the CAdV-WTs in MDCK cells (Figure 3A,B). These data indicated that the recombinant viruses with WT phenotypes could be efficiently and easily rescued by pBAC-CAdV1 or pBAC-CAdV2 clones, verifying our successful establishment of a BAC-based reverse genetics system for CAdVs.
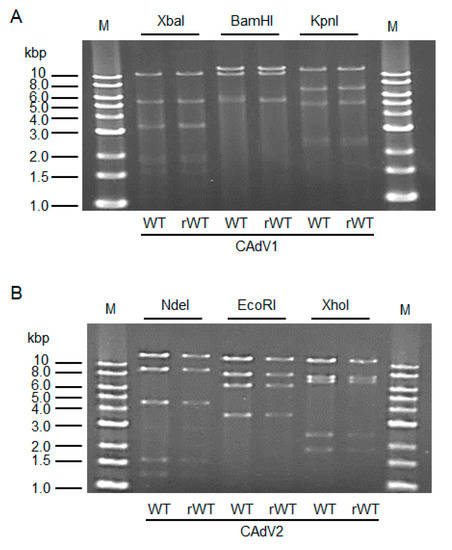
Figure 2. Restriction enzyme analysis of the genomes of wild-type (WT) and recombinant wild-type (rWT) viruses. (A) The genomes of CAdV1-WT and CAdV1-rWT were digested with XbaI, BamHI, and KpnI. The predicted molecular weights of the digested fragments of the viral genomes are 11250, 5551, 3383, 3301, 1955, 1834, 1587, 775, 519, 333, and 45 bp for XbaI; 13554, 10982, 5534, and 463 bp for BamHI; and 12573, 7077, 5110, 2490, 2302, and 981 bp for KpnI. M: 1-kb DNA ladder marker. (B) The genomes of CAdV2-WT and CAdV2-rWT were digested with NdeI, EcoRI, and XhoI. The predicted molecular weights of the digested fragments of the viral genomes are 14820, 9059, 4585, 1603, and 1253 bp for NdeI; 13014, 8192, 6504, and 3610 bp for EcoRI; and 12194, 7688, 7089, 2462, and 1887 bp for XhoI. M: 1-kb DNA ladder marker.
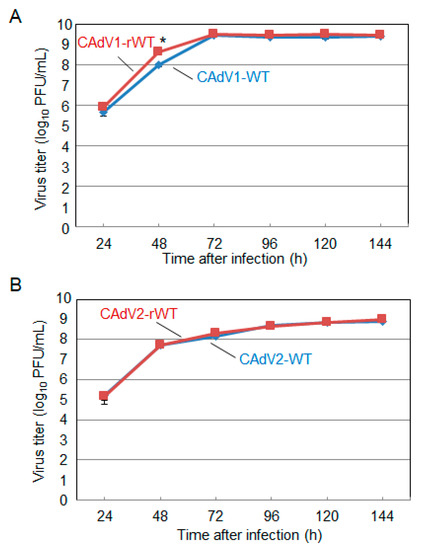
Figure 3. Growth kinetics of the wild-type (WT) and recombinant wild-type (rWT) viruses. MDCK cells were infected with (A) CAdV1-WT or CAdV1-rWT and (B) CAdV2-WT or CAdV2-rWT at a multiplicity of infection (MOI) of 0.01. The culture supernatants were collected at the indicated time points, and the viral titers were determined by plaque assay. The results were reported as the mean titer with standard deviations for three independent experiments. The asterisk indicates a significant difference (p < 0.05 by Student’s t-test).
References
- Walker, D.; Abbondati, E.; Cox, A.L.; Mitchell, G.B.B.; Pizzi, R.; Sharp, C.P.; Philbey, A.W. Infectious canine hepatitis in red foxes (Vulpes vulpes) in wildlife rescue centres in the UK. Vet. Rec. 2016, 178, 421, doi:10.1136/vr.103559.
- García Marín, J.F.; Royo, L.J.; Oleaga, A.; Gayo, E.; Alarcia, O.; Pinto, D.; Martínez, I.Z.; González, P.; Balsera, R.; Marcos, J.L.; et al. Canine adenovirus type 1 (CAdV-1) in free-ranging European brown bear (Ursus arctos arctos): A threat for Cantabrian population? Transbound. Emerg. Dis. 2018, 65, 2049–2056, doi:10.1111/tbed.13013.
- Balboni, A.; Musto, C.; Kaehler, E.; Verin, R.; Caniglia, R.; Fabbri, E.; Carra, E.; Cotti, C.; Battilani, M.; Delogu, M. Genetic characterization of canine adenovirus type 1 detected by real-time polymerase chain reaction in an oral sample of an Italian wolf (Canis lupus). J. Wildl. Dis. 2019, 55, 737–741, doi:10.7589/2018-08-206.
- Ditchfield, J.; Macpherson, L.W.; Zbitnew, A. Association of canine adenovirus (Toronto A 26/61) with an outbreak of laryngotracheitis (“kennel cough”): A preliminary report. Can. Vet. J. 1962, 3, 238–247.
- Decaro, N.; Campolo, M.; Elia, G.; Buonavoglia, D.; Colaianni, M.L.; Lorusso, A.; Mari, V.; Buonavoglia, C. Infectious canine hepatitis: An “old” disease reemerging in Italy. Res. Vet. Sci. 2007, 83, 269–273, doi:10.1016/j.rvsc.2006.11.009.
- Walker, D.; Fee, S.A.; Hartley, G.; Learmount, J.; O’Hagan, M.J.H.; Meredith, A.L.; Bronsvoort, B.M.D.C.; Porphyre, T.; Sharp, C.P.; Philbey, A.W. Serological and molecular epidemiology of canine adenovirus type 1 in red foxes (Vulpes vulpes) in the United Kingdom. Sci. Rep. 2016, 6, 36051, doi:10.1038/srep36051.
- Fueyo, J.; Gomez-Manzano, C.; Alemany, R.; Lee, P.S.; McDonnell, T.J.; Mitlianga, P.; Shi, Y.X.; Levin, V.A.; Yung, W.K.A.; Kyritsis, A.P. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene 2000, 19, 2–12, doi:10.1038/sj.onc.1203251.
- Huang, T.G.; Savontaus, M.J.; Shinozaki, K.; Sauter, B.V.; Woo, S.L.C. Telomerase-dependent oncolytic adenovirus for cancer treatment. Gene Ther. 2003, 10, 1241–1247, doi:10.1038/sj.gt.3301987.
- Kremer, E.J.; Boutin, S.; Chillon, M.; Danos, O. Canine adenovirus vectors: An alternative for adenovirus-mediated gene transfer. J. Virol. 2000, 74, 505–512, doi:10.1128/jvi.74.1.505-512.2000.
- Sánchez, D.; Cesarman-Maus, G.; Amador-Molina, A.; Lizano, M. Oncolytic viruses for canine cancer treatment. Cancers 2018, 10, 404, doi:10.3390/cancers10110404.
- Mizuguchi, H.; Kay, M.A. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 1998, 9, 2577–2583, doi:10.1089/10430349850019418.
- Yuan, Z.G.; Luo, S.J.; Xu, H.J.; Wang, X.H.; Li, J.; Yuan, L.G.; He, L.T.; Zhang, X.X. Generation of E3-deleted canine adenovirus type 2 expressing the Gc glycoprotein of Seoul virus by gene insertion or deletion of related terminal region sequences. J. Gen. Virol. 2010, 91, 1764–1771, doi:10.1099/vir.0.018473-0.
- Yu, Z.; Jiang, Q.; Liu, J.; Guo, D.; Quan, C.; Li, B.; Qu, L. A simplified system for generating recombinant E3-deleted canine adenovirus-2. Plasmid 2015, 77, 1–6, doi:10.1016/j.plasmid.2014.10.005.
- Hu, R.; Zhang, S.; Fooks, A.R.; Yuan, H.; Liu, Y.; Li, H.; Tu, C.; Xia, X.; Xiao, Y. Prevention of rabies virus infection in dogs by a recombinant canine adenovirus type-2 encoding the rabies virus glycoprotein. Microbes Infect. 2006, 8, 1090–1097, doi:10.1016/j.micinf.2005.11.007.
- Klonjkowski, B.; Gilardi-Hebenstreit, P.; Hadchouel, J.; Randrianarison, V.; Boutin, S.; Yeh, P.; Perricaudet, M.; Kremer, E.J. A recombinant E1-deleted canine adenoviral vector capable of transduction and expression of a transgene in human-derived cells and in vivo. Hum. Gene Ther. 1997, 8, 2103–2115, doi:10.1089/hum.1997.8.17-2103.
- Fischer, L.; Tronel, J.P.; Pardo-David, C.; Tanner, P.; Colombet, G.; Minke, J.; Audonnet, J.C. Vaccination of puppies born to immune dams with a canine adenovirus-based vaccine protects against a canine distemper virus challenge. Vaccine 2002, 20, 3485–3497, doi:10.1016/s0264-410x(02)00344-4.
- Chartier, C.; Degryse, E.; Gantzer, M.; Dieterle, A.; Pavirani, A.; Mehtali, M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996, 70, 4805–4810, doi:10.1128/jvi.70.7.4805-4810.1996.
- Van Olphen, A.L.; Mittal, S.K. Generation of infectious genome of bovine adenovirus type 3 by homologous recombination in bacteria. J. Virol. Methods 1999, 77, 125–129, doi:10.1016/S0166-0934(98)00145-1.
- Reddy, P.S.; Idamakanti, N.; Hyun, B.H.; Tikoo, S.K.; Babiuk, L.A. Development of porcine adenovirus-3 as an expression vector. J. Gen. Virol. 1999, 80, 563–570, doi:10.1099/0022-1317-80-3-563.
- Morrison, M.D.; Reid, D.; Onions, D.; Spibey, N.; Nicolson, L. Generation of E3-deleted canine adenoviruses expressing canine parvovirus capsid by homologous recombination in bacteria. Virology 2002, 293, 26–30, doi:10.1006/viro.2001.1262.
- He, T.C.; Zhou, S.; Da Costa, L.T.; Yu, J.; Kinzler, K.W.; Vogelstein, B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 1998, 95, 2509–2514, doi:10.1073/pnas.95.5.2509.
- Ketner, G.; Spencer, F.; Tugendreich, S.; Connelly, C.; Hieter, P. Efficient manupulation of the human adenovirus genome as an infectious yeast aritificial chromosome clone. Proc. Natl. Acad. Sci. USA 1994, 91, 6186–6190, doi:10.1073/pnas.91.13.6186.
