Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Amina Yu and Version 1 by Joanna Grzyb.
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), is one of the best studied enzymes. Rubisco catalyses CO2 assimilation and therefore tt is crucial for photosynthesis, and for all of biosphere’s productivity. There are four isoforms of this enzyme, differing by amino acid sequence composition and quaternary structure. However, there is still a group of organisms, dinoflagellates, single-cell eukaryotes, that are confirmed to possess Rubisco, but no successful purification of the enzyme of such origin, and hence a generation of a crystal structure was reported to date.
- Rubisco
- structure
- photosynthesis
- dinoflagelates
- Symbiodinium sp.
- homohexamer
1. Introduction
Ribulose1,5-bisphosphate carboxylase/oxygenase (Rubisco) is an enzyme employed by plants, algae, cyanobacteria and other autotrophic organisms to incorporate CO2 into organic compounds, thus it is one of the key photosynthetic enzymes. Rubisco catalyses carboxylation reaction, during which it assimilates CO2 and an oxygenation reaction, in which it oxidizes the substrate. In both reactions, the substrate is ribulose-1,5-bisphosphate (RuBP). Due to the fact that Rubisco’s effectiveness of carboxylation is low, and that it also catalyses the unfavourable reaction of photorespiration, it is considered to be a limiting factor of photosynthesis. Consequently, Rubisco is the obvious target for the increase in agricultural production efficiency, and thus it is one of the best studied enzymes for this application [1]. Rubisco consists of at least two catalytic, large subunits (RbcL), and in some cases, of additional regulatory small subunits. To reach catalytic competence, lysine in the active site of Rubisco must first be carboxylated by a non-substrate CO2 molecule, followed by the binding of a Mg2+ ion. This process is called carbamylation and serves to position the substrate RuBP for an efficient electrophilic attack by the second CO2 molecule that will be fixed in the Calvin-Benson cycle (CBB) cycle upon RuBP binding. The active site is closed via two conformational changes in RbcL: loop 6 in the C-terminal domain of RbcL extends over the bound RuBP trapping it underneath; the C-terminal tail of RbcL then stretches across the subunit and pins down loop 6, closing the active site, which results in a closed conformation of Rubisco. Besides RuBP, Rubisco can also bind other molecules like carboxyarabinitol-1,5-bisphosphate (CABP), which is a tight-binding inhibitor of this enzyme, making the active site of carbamylated or decarbamylated Rubisco adopt a closed conformation, and downregulating Rubisco’s activity [2,3,4][2][3][4].
The variation of Rubisco is great due to the huge diversity of organisms that it was found in. Additionally, the different quaternary structures allow distinguishing four different Rubisco forms. Of four known forms, the dinoflagellates form II is the least studied one. Most papers about this form come from the period of 1972–2003 [5,6,7,8,9,10][5][6][7][8][9][10]. Until today all other Rubiscos have been very well studied, while many questions pertaining to the dinoflagellate enzyme are left unanswered. Rubisco from these organisms shows a set of surprising features. Little is known about its catalytic properties, besides the fact that it is highly unstable, however, possesses a much greater specificity factor (SF, defined as the ratio between CO2/O2 activity), than other form II Rubiscos [10,11][10][11]. It is very important to understand the origin of such high SF, as it may help in improving catalytic properties of other Rubiscos. The dinoflagellate Rubisco has been shown to be a form II type enzyme, a homodimer of RbcL (L2), most likely similar to the one from Rhodospirillum rubrum, and is encoded by nuclear-localized genes unlike other known eukaryotic large Rubisco subunits, which are encoded by the plastidic genome. What is more, it is encoded as a triple polyprotein by a diverse gene family that contains introns [6,12][6][12]. Symbiodinium sp. Rubisco expression is photoperiod regulated, but also dependent on its anemone host [13]. Another outstanding fact is that dinoflagellates, although being aerobic photoautotrophs, have a form II Rubisco. This form of Rubisco originates from anaerobic proteobacteria and has a high affinity for O2, and this should lead, under normal circumstances for an aerobic organism, to inefficient CO2 assimilation. Since this is not true, we may suppose that dinoflagellates cells pose a mechanism to cope with the O2 dilemma, e.g., a local CO2 concentrating mechanism (CCM) [7].
This unusual set of features of dinoflagellate Rubisco suggest also unusual evolutionary origin, corresponding to the mysterious evolution of dinoflagellate, with multiple events of endosymbiosis [7]. To further understand it, more data is needed about the enzyme itself.
The main obstacle in obtaining sufficient data is that the dinoflagellate Rubisco is highly unstable. It has been shown that Rubisco from Symbiodinium sp. and A. cartere lost its activity within 30 min following the cell lysis [10], while higher plant or R. rubrum Rubisco is stable for several hours and may be easily isolated [11]. The reason for this venture is not fully understood. It was shown that loss of Symbiodinium sp. Rubisco activity was not due to proteolysis or precipitation. The explanation may be the instability of the L2 dimer or of the higher quaternary structure complex [10]. There might be some specific chaperone proteins involved in stabilising the final oligomer, what is suggested by Rubisco assembly scenario present in other organisms [14,15][14][15]. The existence of chaperones might be deduced from an organism’s genome homology study. However, such is impossible for the dinoflagellate genomes, since they are enormously large (from 1 to 270 Gb, a size that is one-third to 90-fold the size of the human genome), and they have not been fully sequenced so far. Although surely not depicting the whole picture, some chaperones were indeed identified in the Symbiodinium sp. transcriptome [16].
An enzyme’s crystal structure would be helpful in understanding the dinoflagellate Rubisco. No successful effort to solve it was yet carried out, mainly because it is impossible to purify its native form due to the aforementioned. However, tools are available to search for the answers not only in vivo, but also in silico. Such an attempt was successfully used for several proteins, which demonstrated as hard to crystallize [17]. The present paper is an attempt to create a model of a structure of the dinoflagellate Rubisco from Symbiodinium sp. by homology modelling. We utilize known solved structures of form II Rubisco as templates. Then, we show similarities and differences, which we use to build an explanation for the unusual features of dinoflagellate Rubisco. In a basic experiment, we also show that one of the identified elements (an insert forming loop, exclusive for dinoflagellates) may influence Rubisco solubility.
2. Homologues of Form II Rubisco from Rhodospirillum Rubrum among Dinoflagellates
To find the best sequence for further modelling, we used the blastP tool to find homologues of the template R. rubrum Rubisco among dinoflagellates. As mentioned already, this protein is broadly accepted as a model form II Rubisco. The highest scoring entries are listed in Table 1.
Table 1. Highest scoring homologues of Rhodospirillum rubrum Rubisco among dinoflagellates.
| Accession Number | Organism | Query Cover [%] | E Value | Percent Identity [%] |
|---|---|---|---|---|
| Q5ENN5.1 | Heterocapsa triquetra | 97 | 0.0 | 67.67 |
| OLP97682.1 | Symbiodinium microadriaticum | 97 | 0.0 | 65.95 |
| AAO13031.1 | Prorocentrum minimum | 78 | 0.0 | 64.38 |
| Q42813.2 | Lingulodinium polyedra | 97 | 0.0 | 64.24 |
Homologues were searched using the blastP tool with the organism parameter defined to: Dino-flagellates taxid: 2864. Due to the high similarity of sequences between dinoflagellates, only the top 4 are listed in the table. Symbiodinium microadriaticum is listed here, as it is the name of an entry; however, in the hereby text we are using simply Symbiodinium sp., as it is a convention accepted in most of papers pertaining to dinoflagellates.
Heterocapsa triquetra showed the highest similarity of amino acid sequence to the R. rubrum sequence, as described by Query cover (97%, a number that describes how much of the query sequence is covered by the target sequence), E value (0.0, expected value, a number that describes how many times a match by chance in a database of that size is expected; the lower the E value is, the more significant the match) and percent identity (67.67%, a percent of identical amino acids in the same position of the sequence) [18]. The best studied Rubisco from dinoflagellates is the one from Symbiodinium sp., being the second with the highest score. It differs from the first hit by less than 2 in percent identity. Thus, we decided to choose Symbiodinium sp. as a case for further investigations in this paper.
3. Analysis of the Amino Acid Sequence of Dinoflagellate Rubiscos
To compare the primary structure of dinoflagellate Rubisco, we aligned sequences of Rubiscos listed in Table 1 on the R. rubrum template using Clustal OMEGA [14]. This comparison showed differences that might be crucial for further investigation of the eukaryotic form II Rubisco (Figure 1A).


Figure 1. Protein sequence alignment in Clustal OMEGA (A) and a phylogenetic tree of form II Rubiscos from Dinoflagellates constructed based on this alignment (B). Red frames indicate the position of two unique inserts. “*” indicate identical amino acids in all sequences, “:” indicate amino acids which are not identical but have similar properties.
First of all, in our alignment dinoflagellate Rubiscos do not start with a methionine residue (like in R. rubrum), but with a leucine. The lack of an initial codon suggests that there might be a transit peptide encoded at the beginning of the rbcA locus, which encodes rbcL. Rubiscos from dinoflagellates are encoded in the nucleus, and therefore need to be transported into the chloroplasts. It was previously shown that there is an upstream sequence in the rbcA mRNA, with a pattern of conserved residues analogous to Euglena’s Rubisco’s small subunit precursor polyprotein [6]. Aranda and co-workers sequenced and analysed parts of the dinoflagellate genomes and transcriptomes, and identified this upstream sequence of the rbcA locus [8]. The second reason for the lack of methionine is the protein’s encoding as a precursor polyprotein. This means that first result of translation is a longer peptide, bearing a transit peptide, and two or more proteins, which are separated with spacers. This pre-polyprotein trend occurs also in Euglena’s proteome, where, for example, light-harvesting complexes consist as such, and are separated with a deca-peptide spacer [10].
As mentioned previously, more than 67% of the amino acid sequence is identical in aligned proteins. Most of the differences are equally distributed along the compared sequences. The charge distribution is similar; an isoelectric point of Symbiodinium Rubisco is slightly higher than that of R. rubrum enzyme (5.72 vs. 5.60). This is a result of a plus one negative and a minus one positive amino acid in the Symbiodinium sp. sequence. More notable might be the higher amount of cysteine residues in the dinoflagellate Rubisco. In the Symbiodinium sp. sequence, there are 9 such residues, which is almost twice their number (5) in R. rubrum. Notably, only two cysteine residues are conserved between R. rubrum and dinoflagellate Rubiscos (Cys59 and Cys180). Cysteine residues, although not involved directly in Rubisco activity, are known to be responsible for its redox regulation and conformational changes [3,19][3][19]. The importance of cysteine residues was also proven for Arabidopsis thaliana Rubisco; after oxidative inactivation, the enzyme was reactivated by redox treatment [20]. On this basis, we may hypothesise, that the higher content of Cys residues is responsible for possible oxygen-dependent inactivation of Symbiodinium sp. Rubisco upon isolation.
The most significant differences between dinoflagellate and R. Rubrum Rubiscos are the two insertions present in the dinoflagellate RbcL amino acid sequence (Figure 1A, red rectangles). The first insertion contains three negatively charged amino acids in position 413, and the second insertion is made up of eight amino acids in position 425. Both inserts may be treated as one longer, dinoflagellate-specific motif. The possible role of those inserts will be further discussed on the base of constructed models.
4. Conclusions
To conclude, we built a structural model of dinoflagellate Rubisco based on known form II homologs of this enzyme (Fig.2). Dinoflagellates, as mentioned, belong to the Eucaryota, but their Rubisco, classified as type II, is nuclearly encoded in three repeats, differently to other known eukaryotic Rubiscos of type I. This feature may reflect the evolutional history of the Rubisco enzyme, as dinoflagellate Rubisco shows characteristics of both eukaryotic and prokaryotic organisms. It should be kept in mind that this is an in silico study without crystallographic confirmation; however, it comes out with several indications, which may help in further studies. First, we confirmed that the catalytical site of the enzyme is conserved, and therefore is not an explanation for differences noted between dinoflagellate Rubiscos and its homologs from other organisms. Therefore, the experimentally observed loss of activity of isolated dinoflagellate enzyme must be linked to other structural features of the protein.
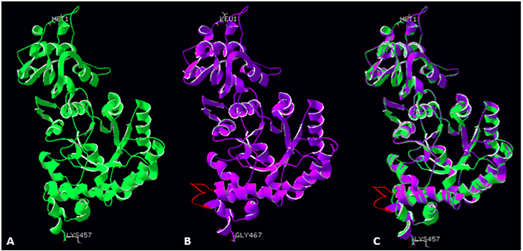

Fig.2.Figure 2 Large subunit monomers from R. palustris (A, green ribbon structure), modelled Symbiodinium sp. Rubisco structure (B, violet ribbon structure), and a superimposition of both structures (C). Red colour indicates a novel loop (insert 425) in the Symbiodinium sp Rubisco structure.
We found, that Rubisco from Symbiodinium sp. has twice as many cysteine residues as the Rubisco from R. rubrum. We postulate that the higher amount of cysteines, which are known to be responsible for redox regulation, might be the cause for high instability of dinoflagellate Rubisco. This observation suggests that the isolation of an active enzyme from a natural source may need additional optimization of redox conditions; the active enzyme expression in a heterological system may also require overcoming of the folding limitations.
Our analysis showed that the dinoflagellatae Rubisco is a hexamer (a trimer of dimers) rather than, as previously suggested, a L2 type enzyme. The indicated hexamer has a more complex structure than a simple dimer. This knowledge might help to obtain a stable purified enzyme, mostly by including chaperone proteins in the process, aiding in formation of a higher oligomer. We may hypothesize that these might be, at least in part, the chaperones alike to those of higher plants; however, it needs further experimental confirmation.
We also show that dinoflagellate Rubiscos contain a novel motif, consisting of a helix extension and a loop. Location of this motif excludes its direct involvement in a catalytical reaction, suggesting rather a role in interaction with an unknown protein partner of possible regulatory function. As a proof of concept, we expressed the Symbiodinium sp. RbcL without the loop, finding the protein solubility to be on a significantly lower level. This loop; therefore, maybe important for the interactions with other proteins, such as a possible unknown regulatory protein as well as chaperones. Again, this makes the dinoflagellate enzyme more similar to the eukaryotic Rubisco due to the similar need for a series of chaperone proteins in order to assemble into an active enzyme. All these findings bring us closer to explaining dinoflagellate Rubisco’s surprising features. Full understanding of Rubisco characteristics will make possible reengineering it to gain a higher yield of CO2 assimilation, what may benefit in higher crop yield and an overall improvement in biosphere CO2 level.
References
- Andersson, I. Catalysis and regulation in Rubisco. J. Exp. Bot. 2008, 59, 1555–1568.
- Duff, A.P.; Andrews, T.J.; Curmi, P.M. The transition between the open and closed states of rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 2000, 298, 903–916.
- Moreno, J.; García-Murria, M.J.; Marín-Navarro, J. Redox modulation of Rubisco conformation and activity through its cysteine residues. J. Exp. Bot. 2008, 59, 1605–1614.
- Tabita, F.R.; Satagopan, S.; Hanson, T.E.; Kreel, N.E.; Scott, S.S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008, 59, 1515–1524.
- Morse, D.; Salois, P.; Markovic, P.; Hastings, J.W. A nuclear-encoded form II RuBisCO in dinoflagellates. Science 1995, 268, 1622–1624.
- Zhang, H.; Lin, S. Complex gene structure of the form II Rubisco in the dinoflagellate Prorocentrum minimum (dinophyceae). J. Phycol. 2003, 39, 1160–1171.
- Palmer, J.D. Rubisco surprises in dinoflagellates. Plant Cell 1996, 8, 343.
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 1–15.
- Rowan, R.; Whitney, S.M.; Fowler, A.; Yellowlees, D. Rubisco in marine symbiotic dinoflagellates: Form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 1996, 8, 539–553.
- Whitney, S.M.; Yellowlees, D. Preliminary investigations into the structure and activity of ribulose bisphosphate carboxylase from two photosynthetic dinoflagellates. J. Phycol. 1995, 31, 138–146.
- Bathellier, C.; Tcherkez, G.; Lorimer, G.H.; Farquhar, G.D. Rubisco is not really so bad. Plant Cell Environ. 2018, 41, 705–716.
- Tessier, L.; Paulus, F.; Keller, M.; Vial, C.; Imbault, P. Structure and expression of Euglena gracilis nuclear rbcS genes encoding the small subunits of the ribulose 1, 5-bisphoshate carboxylase/oxygenase: A novel splicing process for unusual intervening sequences? J. Mol. Biol. 1995, 245, 22–33.
- Mayfield, A.B.; Hsiao, Y.-Y.; Chen, H.-K.; Chen, C.-S. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotechnol. 2014, 16, 371–384.
- Aigner, H.; Wilson, R.; Bracher, A.; Calisse, L.; Bhat, J.; Hartl, F.; Hayer-Hartl, M. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 2017, 358, 1272–1278.
- Hayer-Hartl, M.; Hartl, F.U. Chaperone machineries of rubisco–the most abundant enzyme. Trends Biochem. Sci 2020, 45, 748–763.
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; McDougald, D.; Steinberg, P.D.; Van Oppen, M.J. Sex, scavengers, and chaperones: Transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215.
- Gabruk, M.; Grzyb, J.; Kruk, J.; Mysliwa-Kurdziel, B. Light-dependent and light-independent protochlorophyllide oxidoreductases share similar sequence motifs -in silico studies. Photosynthetica 2012, 50, 529–540.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410.
- García-Murria, M.J.; Sudhani, H.P.; Marín-Navarro, J.; Del Pino, M.M.S.; Moreno, J. Dissecting the individual contribution of conserved cysteines to the redox regulation of RubisCO. Photosynth. Res. 2018, 137, 251–262.
- Tominaga, J.; Takahashi, S.; Sakamoto, A.; Shimada, H. Arabidopsis BSD2 reveals a novel redox regulation of Rubisco physiology in vivo. Plant Signal. Behav. 2020, 15, 1740873.
More
