Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Ron Wang and Version 2 by Ron Wang.
Liver cancer, predominantly hepatocellular carcinoma (HCC), is the third leading cause of cancer-related deaths worldwide. Emerging data highlight the importance of gut homeostasis in the pathogenesis of HCC. Clinical and translational studies revealed the patterns of dysbiosis in HCC patients and their potential role for HCC diagnosis. Research on underlying mechanisms of dysbiosis in HCC development pointed out the direction for improving the treatment and prevention. Despite missing clinical studies, animal models showed that modulation of the gut microbiota by probiotics may become a new way to treat or prevent HCC development.
- hepatocellular carcinoma
- dysbiosis
- microbiota
- probiotics
1. Introduction
Liver cancer, predominantly hepatocellular carcinoma (HCC), is a substantial health burden worldwide. In 2017, with an estimation of 803,400 (753,100 to 856,700) cases, the age-standardized years lived with disability (YLDs) rate increased by 8.1% when compared with that in 2007
[1]
. With a new death of 830,180 cases in 2020, liver cancer represents the third (8.3%) leading cause of cancer-related deaths worldwide
[2]
. Due to the low screening rate in high-risk populations and inadequate sensitivity of the present diagnostic technology (imaging and serum alpha-fetoprotein [AFP] quantification), HCC is usually diagnosed at the late stages, leading to low accessibility of curative therapy and high mortality. Early diagnosis and better prevention and treatment are the goals pursued by doctors and patients together. In terms of diagnostic technology, sensitive and specific biomarkers for early diagnosis of HCC are still lacking. As for prevention and treatment of HCC, apart from etiological treatment of HCC, such as anti-hepatitis B virus (HBV) in HBV-HCC, extra measures are in great need.
Approximately 4 × 10
13
microbial cells spanning ~3 × 10
3
species inhabit the human body. The vast majority (97%) of them are bacteria in the colon, and the remaining include extracolonic bacteria and Archaea and eukaryotes such as fungi
. Gut and liver are closely related, not only anatomically but also functionally. The liver receives blood from the gut through the portal vein, while the gut receives bile from the liver through the bile duct. Blood from the gut brings nutrition, microbial metabolite, and microbe-associated molecular patterns (MAMPs). MAMPs may elicit inflammatory responses by activating pattern recognition receptors (PRRs) in the liver, contributing to the progression of liver diseases and development of HCC. Bile acids, important components in bile, are synthesized from cholesterol in the liver, then metabolized by gut bacteria. They can shape the composition and function of the intestinal microbiota. Mutual interplay of bile acids and gut microbiota regulates many physiological processes
. Emerging data highlight the importance of gut homeostasis in the pathogenesis of HCC. Clinical and translational studies revealed the patterns of dysbiosis in HCC patients, indicating the diagnostic value of the dysbiosis in early diagnosis of HCC. Mechanism research demonstrates that gut microbiota plays an important role in liver tumorigenesis, which suggests the possibility of preventing and treating HCC by modulating gut microbiota.
Although the relationship between gut bacterial microbiota and fibrosis/liver cirrhosis is of importance to understand between gut bacterial microbiota and HCC, previous reviews have discussed this topic in detail
[7][8]. Therefore, in the present review, we only focus on the alteration of gut bacterial microbiota in HCC patients and the underlying mechanisms of dysbiosis in HCC development. Meanwhile, diagnostic value of gut dysbiosis and therapeutic potential by targeting gut dysbiosis in HCC were discussed.
. Therefore, in the present review, we only focus on the alteration of gut bacterial microbiota in HCC patients and the underlying mechanisms of dysbiosis in HCC development. Meanwhile, diagnostic value of gut dysbiosis and therapeutic potential by targeting gut dysbiosis in HCC were discussed.
2. Gut Microbiota Changes in HCC Patients
Gut bacteria dysbiosis in HCC patients has been reported in many countries and regions recently (
Table 1
). Both stool and blood samples possess the value of diagnosing and assessing dysbiosis in HCC patients.
Table 1.
Gut bacteria dysbiosis in HCC patients.
| Patients/Control | Increased Microbiota | Decreased Microbiota | Reference |
|---|---|---|---|
| cirrhotic HCC/cirrhosis | Escherichia coli. | [9] | |
| HCC/NC | Escherichia coli., Enterococcus | Bifidobacterium, Lactobacillus | [10] |
| HCC/cirrhosis HCC/cirrhosis HCC/control |
Actinobacteria Gemmiger, Parabacteroides, Paraprevotella, Clostridium_XVIII Klebsiella and Haemophilus |
Ruminococcus, Oscillibacter, Faecalibacterium, Clostridium IV, and Coprococcus | [11] |
| HCC/NC NBNC-HCC/NC HBV-HCC/NC NBNC-HCC/NC HBV-HCC/NC |
Lactobacillus, Bifidobacterium Proteobacteria Escherichia-Shigella, Enterococcus Faecalibacterium, Ruminococcus, Ruminoclostridium |
Firmicutes Proteobacteria Faecalibacterium, Ruminococcus, Ruminoclostridium |
[12] |
| HCC/NC | Proteobacteria (Enterobacte, Haemophilus) | [13] | |
| NAFLD-HCC/NAFLD-cirrhosis | Bacteroides, Ruminococcaceae | Bifidobacterium | [14] |
| cirrhotic HCC/cirrhosis | Erysipelotrichaceae Odoribacter, Butyricimonas |
Leuconostocaceae Fusobacterium, Lachnospiraceae |
[15] |
| NAFLD-HCC/NAFLD-cirrhosis | Enterobacteriaceae Bacteroides caecimuris, Veillonella parvula, Clostridium bolteae, and Ruminococcus gnavus |
Eubacteriaceae | [16] |
| HCC/NC | Proteobacteria Staphylococcus, Acinetobacter, Klebsiella, Trabulsiella |
Pseudomonas | [17] |
HBV, hepatitis B virus; HCC, hepatocellular carcinoma; NAFLD, nonalcoholic fatty liver disease; NBNC, non-hepatitis B virus non-hepatitis C virus; NC, normal control.
In the early stage, the number of colony-forming units per gram (cfu/g) of wet feces was adopted to analyze the gut bacterial change in HCC patients. Fecal counts of Escherichia coli (E. coli.) increased in 15 cirrhotic HCC patients, when compared to 15 etiology and model for end stage liver disease (MELD) score-matched cirrhosis patients
[9]
. E. coli. and Enterococcus increased, while Bifidobacterium and Lactobacillus significantly decreased in 20 HCC patients vs. 20 normal controls
[10]
.
3. Mechanism Linking Gut Dysbiosis to HCC
3.1. Mechanisms Other Than Bile Acids Dysregulation
More than a decade ago, a mouse model tested the hypothesis that specific intestinal bacteria promote liver cancer in a chemical and viral transgenic mouse model
3.1. Mechanisms Other Than Bile Acids Dysregulation
More than a decade ago, a mouse model tested the hypothesis that specific intestinal bacteria promote liver cancer in a chemical and viral transgenic mouse model[18]
. Underlying mechanisms linking gut dysbiosis to HCC attracted the attention of scientists in the field. So far, leaky gut (a failing gut barrier), bile acids dysregulation, bacterial translocation, endotoxemia and subsequent promotion of liver inflammation, fibrosis, proliferation, and immune suppression have been identified to contribute to the development of HCC in the setting of chronic liver diseases (
Figure 1
). The concept of the gut–liver axis, bidirectional relationship between the gut and its microbiota, provides the possibility of preventing and treating HCC by targeting gut and its microbiota
[16]
.
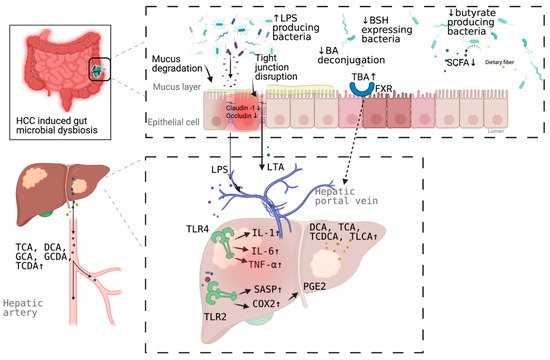

Figure 1.
Schematic representation of the mechanism of the promotion and progresssion of HCC by gut microbiota. BA, bile acid; TBA, total bile acid; LPS; Lipopolysaccharides; BSH, bile salt hydrolase; LTA, Lipoteichoic acid; SCFA, short chain fatty acid; DCA, deoxycholic acid; TCA, taurocholic acid; GCA, glycocholic acid; GCDA, glycochenodeoxycholic acid; TCDA, taurochenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; TLCA, taurolithocholic acid (TLCA); PGE2, prostaglandin E2; COX2, cyclooxygenase-2; SASP, senescence associated secretory phenotype; TLR, toll-like receptor, FXR, farnesoid X receptor. Figure created with BioRender.com (San Francisco, CA, USA).
3.2. Bile Acids Dysregulation in HCC
Emerging evidence indicates the association between bile acid–bacterial microbiota crosstalk and the development of HCC. Bile acids, synthesized from cholesterol in the liver, are metabolized by gut bacteria and subsequently sensed by two major sensing receptors, farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (GPBAR1) (transmembrane G-protein-coupled receptor 5, [TGR5]). Bile acids pools comprise a variety of species of amphipathic acidic steroids and have both protective and pathogenic roles in liver diseases. Hydrophilic bile acids, such as ursodeoxycholic acid (UDCA), and its taurine-conjugated form tauroursodeoxycholic acid (TUDCA), show profound cytoprotective properties[5]
, while excessive production of hydrophobic bile acids is cytotoxic and promotes hepatocyte injury
[19]
.
FXR, a nuclear receptor, activation is involved in regulating antibacterial defense in the small intestine
[20]
, preventing chemically induced intestinal inflammation
[21]
and modulating liver regeneration
[22]
. FXR and epidermal growth factor receptor (EGFR) signaling is involved in regulating intestinal cell proliferation by bile acids
[23]
. In addition, FXR and small heterodimer partner (SHP) can regulate protein N-glycan modifications in the liver
[24]
. TGR5, a plasma membrane receptor, is expressed in sinusoidal endothelial cells, Kupffer cells, cholangiocytes and activated hepatic stellate cells, modulating microcirculation, inflammation, regeneration, biliary secretion, and gallbladder filling
[25]
.
Multiple genera of the gut microbiota are involved in bile acid metabolism, including Bacteroides, Clostridium, Lactobacillus, Bifidobacterium, and Listeria in bile acid deconjugation; Bacteroides, Eubacterium, Clostridium, Escherichia, Egghertella, Eubacterium, Peptostreptococcus, and Ruminococcus in oxidation and epimerization of hydroxyl groups at C3, C7, and C12; Clostridium and Eubacterium in 7-dehydroxylation; Bacteroides, Eubacterium, and Lactobacillus in esterification; and Clostridium, Fusobacterium, Peptococcus, and Pesudomonas in desulfation
[26]
. Dysbiosis of gut microbiota can affect the bile acids homeostasis theoretically. Indeed, clinical studies and animal models demonstrated the dysbacteriosis of some of these above-mentioned bacteria, dysregulation of bile acids in multiple samples (liver tissue, serum, feces, and urine), and the association between the above two abnormalities
.
4. Microbial Dysbiosis in HCC Diagnosis
Both fecal and circulating microbial dysbiosis have the potential value in diagnosis of HCC (
Table 2
).
Table 2.
Diagnostic value of microbiota and metabolites in HCC.
| Microbiota | 1 | Patients/Control | AUC | 95% CI | Sensitivity | Specificity | Reference |
|---|---|---|---|---|---|---|---|
| Escherichia coli | HCC/cirrhosis | 0.742 | 0.564–0.920 | 66.7% | 73.3% | [9] | |
| 30 OTUs markers | HCC/non-HCC | 0.806 | 0.745–0.868 | - | - | [11] | |
| Enterococcus | Cirrhotic HCC/cirrhosis | 0.868 | -NA | 95.8% | 69.2% | [34] | |
| Enterococcus | Non-cirrhotic HCC/cirrhosis | 0.899 | NA | 100% | 78.3% | ||
| Limnobacter | Non-cirrhotic HCC/cirrhosis | 0.858 | NA | 62.5% | 91.3% | ||
| Phyllobacterium | Non-cirrhotic HCC/cirrhosis | 0.868 | NA | 75.0% | 91.3% | ||
| 5 OTUs markers (serum) | HCC/control | 0.879 | NA | 72.9% | 85.0% | [17] | |
| Phe-Trp + GCA (serum) | HCC/cirrhosis | 0.807 | 0.753–0.861 | 92.1% | 52.8% | [28] | |
| Phe-Trp + GCA +AFP (serum) | HCC/cirrhosis | 0.826 | 0.774–0.877 | 77.9% | 76.4% | ||
| CDCA + LPC 20:5 + succinyladenosine + uridine (serum) | HCC/cirrhosis | 0.938 | - | 93.3% | 86.7% | [29] |
1 feces sample is used if not specified. AFP, alpha-fetoprotein; AUC, area under the curve; CDCA, chenodeoxycholic acid; CI, confidence interval; GCA; glycocholate; HCC, hepatocellular carcinoma; LPC, lysophosphatidylcholine; NA, not available; OTU, operational taxonomic unit; Phe-Trp, phenylalanyl-tryptophan. -NA: failed to find out the 95%CI from the paper.
feces sample is used if not specified. AFP, alpha-fetoprotein; AUC, area under the curve; CDCA, chenodeoxycholic acid; CI, confidence interval; GCA; glycocholate; HCC, hepatocellular carcinoma; LPC, lysophosphatidylcholine; NA, not available; OTU, operational taxonomic unit; Phe-Trp, phenylalanyl-tryptophan. -NA: failed to find out the 95%CI from the paper.
5. Targeting Microbial Dysbiosis in HCC Treatment and Prevention
The clear role microbial dysbiosis in the development of HCC, offers multiple pathways and targets for HCC treatment and prevention theoretically.
For example, PGE2 and its receptor may be novel therapeutic targets for noncirrhotic NASH-associated HCC
[35]
. Blocking DCA production or reducing gut bacteria efficiently prevents HCC development in obese mice
[36]
. Gut sterilization can prevent HCC in a mouse model, suggesting that the intestinal microbiota and TLR4 represent therapeutic targets for HCC prevention in advanced liver disease. TLR antagonists can block the propagation of downstream cytokine release
. Reduction of HCC development by modulating gut microbiota was showed in animal models
. Antibiotics can be used to eliminate disease-promoting bacteria and decrease release of MAMPs and metabolites from a leaky gut. FXR agonists can modulate various downstream immune-related pathways.
Fecal microbiota transplantation (FMT) is the transfer of stool from a healthy donor into the gastrointestinal tract, aiming to gain a therapeutic benefit by changing or normalizing the recipient’s gut microbiota directly. FMT has been approved for treating recurrent and refractory Clostridium difficile infection (CDI) by the United States Food and Drug Administration. In the field of treating liver diseases, FMT can improve neurocognitive function and reduce the readmission of patients with hepatic encephalopathy (HE), despite the small scale of study and absence of long-term follow-up
[40]
. What is more gratifying is that microbiota originating from donors was found in human recipients one year after FMT
[40]
. However, clinical study regarding FMT in the treatment and prevention of HCC is still missing.
Probiotics can be used to restore normal microbiota composition, suppress the growth of pathogenic microorganisms, and interact with the mucosal system, which affects the systemic immunity. Administration of a commercial probiotic compound VSL#3 (VSL Pharmaceuticals, Fort Lauderdale, FL, USA) dramatically suppressed penicillin-increased HCC formation in rats
[41]
. A mouse model demonstrated that the efficacy of a novel probiotic mixture (Prohep) slows down the tumor growth significantly and reduces the tumor size and weight by 40% compared with the control
[42]
. Notably, Prohep limits tumor growth by reducing angiogenesis, and so forth lead to hypoxia-induced cell death in tumor. This indicates that combining Prohep with drugs of other mechanisms, such as immunotherapy, may play a synergistic therapeutic effect.
Given the BA-bacterial microbiota crosstalk in the development of HCC, restoring bile acids homeostasis by modulating gut microbiota or targeting directly bile acids may be effective strategies on preventing and treating HCC. Treatment with antibiotics dramatically reduced the accumulation of secondary bile acids and significantly suppressed tumor developments in the HFD mouse model
[43]
. An obese mouse model showed that blocking DCA production or reducing gut bacteria by oral antibiotic caused a marked reduction of HCC development in obese mice
[36]
. Treatment with antibiotics significantly attenuated liver pathology and suppressed tumor development in a new class NASH-inducing HFD mouse model
[43]
. In addition, oral administration of cholestyramine, bile acid sequestrant to enhance intestinal excretion of hydrophobic bile acids, significantly prevent HCC in a mouse model
[33]
. Depleting Gram-positive bacteria by vancomycin treatment can induce hepatic NKT cell accumulation and suppress liver tumor growth in multiple mouse models, while feeding secondary bile acids or colonization of bile acid-metabolizing bacteria can reverse both NKT cell accumulation and inhibition of liver tumor growth in mice
[44].
Together, targeting microbial dysbiosis to treat and prevent HCC seems promising. However, there is no clinical data in this regard currently.
.
Together, targeting microbial dysbiosis to treat and prevent HCC seems promising. However, there is no clinical data in this regard currently.
References
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021.
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371.
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533.
- Amaral, J.D.; Viana, R.J.; Ramalho, R.M.; Steer, C.J.; Rodrigues, C.M. Bile acids: Regulation of apoptosis by ursodeoxycholic acid. J. Lipid Res. 2009, 50, 1721–1734.
- Gruner, N.; Mattner, J. Bile acids and microbiota: Multifaceted and versatile regulators of the liver-gut axis. Int. J. Mol. Sci. 2021, 22, 1397.
- Li, R.; Mao, Z.; Ye, X.; Zuo, T. Human gut microbiome and liver diseases: From correlation to causation. Microorganisms 2021, 9, 1017.
- Trebicka, J.; Macnaughtan, J.; Schnabl, B.; Shawcross, D.L.; Bajaj, J.S. The microbiota in cirrhosis and its role in hepatic decompensation. J. Hepatol. 2021, 75 (Suppl. 1), S67–S81.
- Grat, M.; Wronka, K.M.; Krasnodebski, M.; Masior, L.; Lewandowski, Z.; Kosinska, I.; Grat, K.; Stypulkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant. Proc. 2016, 48, 1687–1691.
- Xin, H.; Li, X.; Sun, R.; Meng, Y.; Yu, Q.; Hao, Y. Endotoxin and intestinal microflora in patients with hepatocellular carcinoma. Chin. J. Gen. Surg. 2019, 34, 686–688.
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023.
- Liu, Q.; Li, F.; Zhuang, Y.; Xu, J.; Wang, J.; Mao, X.; Zhang, Y.; Liu, X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019, 11, 1.
- Ni, J.; Huang, R.; Zhou, H.; Xu, X.; Li, Y.; Cao, P.; Zhong, K.; Ge, M.; Chen, X.; Hou, B.; et al. Analysis of the relationship between the degree of dysbiosis in gut microbiota and prognosis at different stages of primary hepatocellular carcinoma. Front. Microbiol. 2019, 10, 1458.
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019, 69, 107–120.
- Pinero, F.; Vazquez, M.; Bare, P.; Rohr, C.; Mendizabal, M.; Sciara, M.; Alonso, C.; Fay, F.; Silva, M. A different gut microbiome linked to inflammation found in cirrhotic patients with and without hepatocellular carcinoma. Ann. Hepatol. 2019, 18, 480–487.
- Behary, J.; Amorim, N.; Jiang, X.T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021, 12, 187.
- Cho, E.J.; Leem, S.; Kim, S.A.; Yang, J.; Lee, Y.B.; Kim, S.S.; Cheong, J.Y.; Cho, S.W.; Kim, J.W.; Kim, S.M.; et al. Circulating microbiota-based metagenomic signature for detection of hepatocellular carcinoma. Sci. Rep. 2019, 9, 7536.
- Fox, J.G.; Feng, Y.; Theve, E.J.; Raczynski, A.R.; Fiala, J.L.; Doernte, A.L.; Williams, M.; McFaline, J.L.; Essigmann, J.M.; Schauer, D.B.; et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 2010, 59, 88–97.
- Marin, J.J.; Macias, R.I.; Briz, O.; Banales, J.M.; Monte, M.J. Bile acids in physiology, pathology and pharmacology. Curr. Drug Metab. 2015, 17, 4–29.
- Inagaki, T.; Moschetta, A.; Lee, Y.K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925.
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.; Renooij, W.; Murzilli, S.; Klomp, L.W.; Siersema, P.D.; Schipper, M.E.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472.
- Li, G.; Guo, L.G. Farnesoid X receptor, the bile acid sensing nuclear receptor, in liver regeneration. Acta Pharm. Sin. B 2015, 5, 93–98.
- Dossa, A.Y.; Escobar, O.; Golden, J.; Frey, M.R.; Ford, H.R.; Gayer, C.P. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G81–G92.
- Mathur, B.; Shajahan, A.; Arif, W.; Chen, Q.; Hand, N.J.; Abramowitz, L.K.; Schoonjans, K.; Rader, D.J.; Kalsotra, A.; Hanover, J.A.; et al. Nuclear receptors FXR and SHP regulate protein N-glycan modifications in the liver. Sci. Adv. 2021, 7.
- Keitel, V.; Haussinger, D. Role of TGR5 (GPBAR1) in liver disease. Semin. Liver Dis. 2018, 38, 333–339.
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128.
- Gao, L.; Lv, G.; Li, R.; Liu, W.T.; Zong, C.; Ye, F.; Li, X.Y.; Yang, X.; Jiang, J.H.; Hou, X.J.; et al. Glycochenodeoxycholate promotes hepatocellular carcinoma invasion and migration by AMPK/mTOR dependent autophagy activation. Cancer Lett. 2019, 454, 215–223.
- Luo, P.; Yin, P.; Hua, R.; Tan, Y.; Li, Z.; Qiu, G.; Yin, Z.; Xie, X.; Wang, X.; Chen, W.; et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 2018, 67, 662–675.
- Han, J.; Qin, W.X.; Li, Z.L.; Xu, A.J.; Xing, H.; Wu, H.; Zhang, H.; Wang, M.D.; Li, C.; Liang, L.; et al. Tissue and serum metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin. Chim. Acta 2019, 488, 68–75.
- Sydor, S.; Best, J.; Messerschmidt, I.; Manka, P.; Vilchez-Vargas, R.; Brodesser, S.; Lucas, C.; Wegehaupt, A.; Wenning, C.; Assmuth, S.; et al. Altered microbiota diversity and bile acid signaling in cirrhotic and noncirrhotic NASH-HCC. Clin. Transl. Gastroenterol. 2020, 11, e00131.
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X.; et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell Proteom. 2011, 10, M110.004945.
- Xiao, J.F.; Varghese, R.S.; Zhou, B.; Nezami Ranjbar, M.R.; Zhao, Y.; Tsai, T.H.; Di Poto, C.; Wang, J.; Goerlitz, D.; Luo, Y.; et al. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J. Proteome Res. 2012, 11, 5914–5923.
- Xie, G.; Wang, X.; Huang, F.; Zhao, A.; Chen, W.; Yan, J.; Zhang, Y.; Lei, S.; Ge, K.; Zheng, X.; et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int. J. Cancer 2016, 139, 1764–1775.
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250.
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut microbiota promotes obesity-associated liver cancer through PGE2-mediated suppression of antitumor immunity. Cancer Discov. 2017, 7, 522–538.
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101.
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516.
- Darnaud, M.; Faivre, J.; Moniaux, N. Targeting gut flora to prevent progression of hepatocellular carcinoma. J. Hepatol. 2013, 58, 385–387.
- Yu, L.X.; Yan, H.X.; Liu, Q.; Yang, W.; Wu, H.P.; Dong, W.; Tang, L.; Lin, Y.; He, Y.Q.; Zou, S.S.; et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 2010, 52, 1322–1333.
- Madsen, M.; Kimer, N.; Bendtsen, F.; Petersen, A.M. Fecal microbiota transplantation in hepatic encephalopathy: A systematic review. Scand. J. Gastroenterol. 2021, 56, 560–569.
- Zhang, H.L.; Yu, L.X.; Yang, W.; Tang, L.; Lin, Y.; Wu, H.; Zhai, B.; Tan, Y.X.; Shan, L.; Liu, Q.; et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J. Hepatol. 2012, 57, 803–812.
- Li, J.; Sung, C.Y.; Lee, N.; Ni, Y.; Pihlajamaki, J.; Panagiotou, G.; El-Nezami, H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315.
- Yamada, S.; Takashina, Y.; Watanabe, M.; Nagamine, R.; Saito, Y.; Kamada, N.; Saito, H. Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget 2018, 9, 9925–9939.
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360.
More
