The application of metabolomics in searching for the mechanisms of the polycystic ovary syndrome (PCOS) pathophysiology gives a promising insight into the research on PCOS. There is a need to investigate the metabolic pathways, which could be involved in the pathophysiology of PCOS and to find the metabolic markers of this disorder. Metabolomics is a valuable and rapidly expanding tool, enabling the discovery of novel metabolites, which may be the potential biomarkers of metabolic and endocrine disorders.
The application of metabolomics in searching for the mechanisms of the polycystic ovary syndrome (PCOS) pathophysiology gives a promising insight into the research on PCOS. There is a need to
investigate the metabolic pathways, which could be involved in the pathophysiology of PCOS and to
find the metabolic markers of this disorder. Metabolomics is a valuable and rapidly expanding tool, enabling the discovery of novel metabolites, which may be the potential biomarkers of metabolic and endocrine disorders.
- metabolomics
- polycystic ovary syndrome (PCOS)
- metabolites
- biomarkers
- mass spectrometry
1. IDefintroductition
Polycystic ovary syndrome (PCOS) is a complex endocrinopathy, which affects more than 10% of women of reproductive age
Polycystic ovary syndrome (PCOS) is a complex endocrinopathy, which affects more than 10% of women of reproductive age[1]
. It is the main cause of female infertility due to oligo- or anovulation. Despite such a high incidence, the pathogenesis of PCOS is still unexplained. Some studies suggest that it is due to the genetic factors associated with ovarian steroidogenesis[2]
. According to the Androgen Excess and PCOS Society (AE&PCOS), the diagnosis of PCOS should be based on the presence of clinical and/or biochemical hyperandrogenism (HA) and the ovarian dysfunction defined as menstrual abnormalities (anovulatory oligomenorrhea (AnO)) or/and the presence of the polycystic ovary morphology (PCOM) in the transvaginal ultrasound (TV-US)[3]
. These criteria yield three separate PCOS phenotypes: A, B, and C. Phenotype A includes all the three features (HA, AnO, and PCOM) whereas phenotype B and C only two (HA and AnO or HA and PCOM, respectively). However, regarding Rotterdam criteria, the fourth phenotype (D) was separated to comprise AnO and PCOM presence. The clinical symptoms of hyperandrogenism include hirsutism (present in 60% of women), androgenic alopecia, and acne, which negatively affect women’s psyche, their femininity and lead to low self-esteem and depression[4]
. In addition to the reproductive and endocrine dysfunction, PCOS is characterized by intrinsic insulin resistance (IR), which lead to the development of the metabolic syndrome (MetS) and its consequences such as disturbed carbohydrate metabolism and type 2 diabetes mellitus (T2DM). Most common clinical manifestation in PCOS is abdominal obesity, which is involved in the development of dyslipidemia, arterial hypertension (AH), as well as non-alcoholic fatty liver disease (NAFLD) . These in turn lead to the development of cardiovascular disease (CVD), which still remains the main cause of death among women[9]
. The clinical picture of this complex endocrinopathy was presented in . Therefore, the treatment of PCOS focuses not only on the symptoms of hyperandrogenism and infertility, but also on improving IR and its metabolic consequences[10]
. Thus, there is a need for a better understanding of the pathomechanisms of this complex disorder through the identification of potential biomarkers with the use of new, non-invasive and specific methods. In recent years, one of the developing scientific approaches is metabolomics[11].
.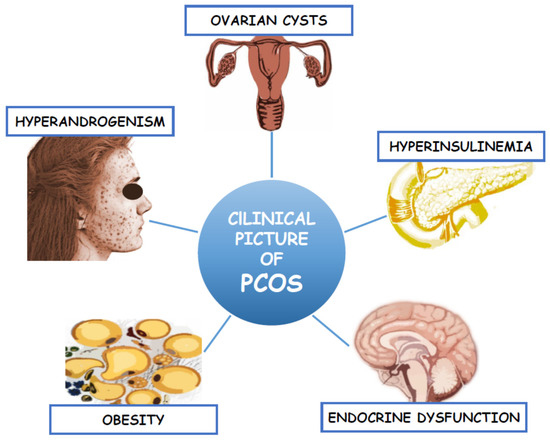
Figure 1. Clinical picture of Polycystic Ovary Syndrome.
2. Metabolomic Approach in Studying the Pathogenesis of Polycystic Ovary Syndrome
Among “omics” techniques, metabolomics plays an important role in studying the potential mechanisms responsible for the development of PCOS. Metabolomics allows to identify and quantify small molecules, which occur in all living organisms
Among “omics” techniques, metabolomics plays an important role in studying the potential mechanisms responsible for the development of PCOS. Metabolomics allows to identify and quantify small molecules, which occur in all living organisms[12]
. The set of all human metabolites that have been identified so far is stored in the Human Metabolome Database (HMDB). Each year, the number of identified metabolites grows. Few years ago, about 41,000 metabolites were found, but now this database contains over 114,190 compounds. Among them, the following groups can be found: amino acids, lipids, peptides, vitamins, organic acids and both endo- and exogenous carbohydrates. Therefore, metabolomics serves as a valuable source of information. The metabolome indicates not only a genetically determined phenotype, but also points to the differences determined by other factors, such as age, diet, or physical activity. The application of metabolomics enables monitoring of the state of an organism and provides information on the compounds formed as a result of many biochemical processes. Any disturbances occurring in a living organism cause changes to the qualitative and quantitative profile of the metabolites. The metabolome describes both the physiological and pathological state of the organism. For this reason, it is known to be an attractive approach, compared to genomics and proteomics, which only suggest the presence of metabolic derangements that occur in the organism . Due to this fact, the use of metabolomics in studying the pathophysiology of PCOS allows to monitor even the smallest biochemical changes in this endocrinopathy and therefore, may help in its diagnosis[17].
.Among the many analytical techniques, chromatography coupled with mass spectrometry (MS) seems to be the “gold technique”. While chromatography allows for the separation of metabolites present in complex, biological samples, mass spectrometry provides specific information about the chemical structure of the compounds, such as characteristic fragmentation ions, accurate mass, and isotope distribution pattern utilized for the identification of metabolites. MS characterizes very high selectivity and sensitivity that allows to detect and measure trace amounts of metabolites
Among the many analytical techniques, chromatography coupled with mass spectrometry (MS) seems to be the “gold technique”. While chromatography allows for the separation of metabolites present in complex, biological samples, mass spectrometry provides specific information about the chemical structure of the compounds, such as characteristic fragmentation ions, accurate mass, and isotope distribution pattern utilized for the identification of metabolites. MS characterizes very high selectivity and sensitivity that allows to detect and measure trace amounts of metabolites[18]
. The combination of MS with gas chromatography (GC-MS) and liquid chromatography (LC-MS) enables to analyse complex biological samples broadly used in metabolomics. GC-MS is suitable for volatile and non-volatile compounds, which require a derivatization step, but first of all thermally stable analytes. The LC-MS technique is widely used for targeted and non-targeted metabolomic analysis and allows to qualify and identify more polar compounds[19]
. Nuclear magnetic resonance (NMR), despite its lower sensitivity than MS, allows to analyse metabolites that are difficult to ionize or require derivative reaction for MS and identify compounds with the same masses[20]. A combination of these complementary techniques enables to analyse a broader array of metabolites and offers more certain results than their separate use.
. A combination of these complementary techniques enables to analyse a broader array of metabolites and offers more certain results than their separate use.3. Matrices for Metabolomic Studies
The application of metabolomics allows the use of several matrices such as tissue and body fluids (i.e., plasma, serum, saliva, follicular fluid, semen). The choice of the matrices is associated with the aim of the conducted study as well as the characteristics of the studied disorder. Ovarian tissue can also be used; however, sampling is invasive and problematic. It is usually obtained during laparoscopic wedge resection surgery. For this reason, the use of ovarian tissue in studying the pathophysiology of PCOS is not very common. The matrices widely used in metabolomic studies associated with PCOS are plasma and urine. Serum and urine samples are more common, because they are easily collectible and simple to prepare. On comparing the significantly altered metabolites, it can be observed that the results obtained for both matrices do not completely overlap. The new alternative matrix is follicular fluid, which is innovative in case of PCOS research, especially in terms of oocytes maturation and their quality
The application of metabolomics allows the use of several matrices such as tissue and body fluids (i.e., plasma, serum, saliva, follicular fluid, semen). The choice of the matrices is associated with the aim of the conducted study as well as the characteristics of the studied disorder. Ovarian tissue can also be used; however, sampling is invasive and problematic. It is usually obtained during laparoscopic wedge resection surgery. For this reason, the use of ovarian tissue in studying the pathophysiology of PCOS is not very common. The matrices widely used in metabolomic studies associated with PCOS are plasma and urine. Serum and urine samples are more common, because they are easily collectible and simple to prepare. On comparing the significantly altered metabolites, it can be observed that the results obtained for both matrices do not completely overlap. The new alternative matrix is follicular fluid, which is innovative in case of PCOS research, especially in terms of oocytes maturation and their quality .References
- Teimuraz Apridonidze; P. A. Essah; Maria J. Iuorno; John E. Nestler; Prevalence and Characteristics of the Metabolic Syndrome in Women with Polycystic Ovary Syndrome. The Journal of Clinical Endocrinology & Metabolism 2005, 90, 1929-1935, 10.1210/jc.2004-1045.
- Raiane P. Crespo; Tania A. S. S. Bachega; Berenice Bilharinho Mendonca; Larissa G. Gomes; An update of genetic basis of PCOS pathogenesis. Archives of Endocrinology and Metabolism 2018, 62, 352-361, 10.20945/2359-3997000000049.
- Robert L. Rosenfield; The Diagnosis of Polycystic Ovary Syndrome in Adolescents. Pediatrics 2015, 136, 1154-1165, 10.1542/peds.2015-1430.
- Fatemeh Nasiri Amiri; Fahimeh Ramezani Tehrani; Masoumeh Simbar; Ali Montazeri; Reza Ali Mohammadpour Thamtan; The Experience of Women Affected by Polycystic Ovary Syndrome: A Qualitative Study From Iran. International Journal of Endocrinology and Metabolism 2014, 12, e13612, 10.5812/ijem.13612.
- Ana Luiza Lunardi Rocha; L. C. Faria; T. C. M. Guimarães; G. V. Moreira; A. L. Cândido; Cláudia Alves Couto; Fernando M. Reis; Non-alcoholic fatty liver disease in women with polycystic ovary syndrome: systematic review and meta-analysis. Journal of Endocrinological Investigation 2017, 40, 1279-1288, 10.1007/s40618-017-0708-9.
- Enrico Carmina; Salvo Bucchieri; Antonella Esposito; Antonio Del Puente; Pasquale Mansueto; Francesco Orio; Gaetana Di Fede; Giovambattista Rini; Abdominal Fat Quantity and Distribution in Women with Polycystic Ovary Syndrome and Extent of Its Relation to Insulin Resistance. The Journal of Clinical Endocrinology & Metabolism 2007, 92, 2500-2505, 10.1210/jc.2006-2725.
- Pallavi Shukla; Srabani Mukherjee; Mitochondrial dysfunction: An emerging link in the pathophysiology of polycystic ovary syndrome. Mitochondrion 2020, 52, 24-39, 10.1016/j.mito.2020.02.006.
- Qi Liu; Yuan-Jie Xie; Li-Hua Qu; Meng-Xia Zhang; Zhong-Cheng Mo; Dyslipidemia involvement in the development of polycystic ovary syndrome.. Taiwanese Journal of Obstetrics and Gynecology 2019, 58, 447-453, 10.1016/j.tjog.2019.05.003.
- Georgios Papadakis; Evanthia Diamanti-Kandarakis; Olga Papalou; Andromachi Vryonidou; Evanthia Diamanti-Kandarakis; Is cardiovascular risk in women with PCOS a real risk? Current insights.. Minerva Endocrinologica 2017, 42, 340-355, 10.23736/S0391-1977.17.02609-8.
- Ricardo Azziz; Enrico Carmina; Zi-Jiang Chen; Andrea Dunaif; Joop S. E. Laven; Richard S. Legro; Daria V. Lizneva; Barbara Natterson-Horowtiz; Helena J Teede; Bulent O. Yildiz; et al. Polycystic ovary syndrome. Nature Reviews Disease Primers 2016, 2, 16057, 10.1038/nrdp.2016.57.
- Mora Murri; María Insenser; Héctor F. Escobar-Morreale; Metabolomics in polycystic ovary syndrome. Clinica Chimica Acta 2014, 429, 181-188, 10.1016/j.cca.2013.12.018.
- Shi-Kai Yan; Run-Hui Liu; Hui-Zi Jin; Xin-Ru Liu; Ji Ye; Lei Shan; Wei-Dong Zhang; “Omics” in pharmaceutical research: overview, applications, challenges, and future perspectives. Chinese Journal of Natural Medicines 2015, 13, 3-21, 10.1016/s1875-5364(15)60002-4.
- Evanthia Diamanti-Kandarakis; Christina Piperi; Georgia Argyrakopoulou; Jiovanna Spina; Lambrini Papanastasiou; Angeliki Bergiele; Dimitrios Panidis; Polycystic Ovary Syndrome: The influence of environmental and genetic factors. Hormones 2006, 5, 17-34, 10.14310/horm.2002.11165.
- John C Lindon; Elaine Holmes; Mary E. Bollard; Elizabeth G. Stanley; Jeremy K. Nicholson; Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers 2004, 9, 1-31, 10.1080/13547500410001668379.
- Manuel Luque-Ramírez; José Luis San Millán; Héctor F. Escobar-Morreale; Genomic variants in polycystic ovary syndrome. Clinica Chimica Acta 2006, 366, 14-26, 10.1016/j.cca.2005.10.017.
- W Atiomo; S Khalid; S Parameshweran; M Houda; Robert Layfield; Proteomic biomarkers for the diagnosis and risk stratification of polycystic ovary syndrome: a systematic review. BJOG: An International Journal of Obstetrics & Gynaecology 2008, 116, 137-143, 10.1111/j.1471-0528.2008.02041.x.
- María G. Barderas; Carlos M. Laborde; Maria Posada-Ayala; Fernando De La Cuesta; Irene Zubiri; Fernando Vivanco; Gloria Alvarez-Llamas; Metabolomic Profiling for Identification of Novel Potential Biomarkers in Cardiovascular Diseases. Journal of Biomedicine and Biotechnology 2011, 2011, 1-9, 10.1155/2011/790132.
- Zhentian Lei; David V. Huhman; Lloyd W. Sumner; Mass Spectrometry Strategies in Metabolomics. Journal of Biological Chemistry 2011, 286, 25435-25442, 10.1074/jbc.r111.238691.
- Silas G. Villas-Bôas; Sandrine Mas; Mats Åkesson; Jørn Smedsgaard; J. Nielsen; Mass spectrometry in metabolome analysis. Mass Spectrometry Reviews 2005, 24, 613-646, 10.1002/mas.20032.
- John L. Markley; Rafael Brüschweiler; Arthur S. Edison; Hamid R. Eghbalnia; Robert Powers; Daniel Raftery; David S. Wishart; The future of NMR-based metabolomics. Current Opinion in Biotechnology 2017, 43, 34-40, 10.1016/j.copbio.2016.08.001.
- Xiang Ma; Lu Fan; Yan Meng; Zheng Hou; Yun-Dong Mao; Wei Wang; Wei Ding; Jiayin Liu; Proteomic analysis of human ovaries from normal and polycystic ovarian syndrome. Molecular Human Reproduction 2007, 13, 527-535, 10.1093/molehr/gam036.
- B. Matharoo-Ball; C. Hughes; Lee Lancashire; D. Tooth; Graham Ball; Colin S. Creaser; M. Elgasim; R. Rees; Rob Layfield; William Atiomo; et al. Characterization of Biomarkers in Polycystic Ovary Syndrome (PCOS) Using Multiple Distinct Proteomic Platforms†. Journal of Proteome Research 2007, 6, 3321-3328, 10.1021/pr070124b.
- Charles W Schmidt; Metabolomics: what's happening downstream of DNA.. Environmental Health Perspectives 2004, 112, A410-A415, 10.1289/ehp.112-a410.
