Micro RNAs (miRNAs) are a class of small non-coding RNAs that have a crucial role in cellular processes such as differentiation, proliferation, migration, and apoptosis. miRNAs may act as oncogenes or tumor suppressors; therefore, they prevent or promote tumorigenesis, and abnormal expression has been reported in many malignancies. The role of miRNA in leukemia pathogenesis is still emerging, but several studies have suggested using miRNA expression profiles as biomarkers for diagnosis, prognosis, and response to therapy in leukemia.
- circulating miRNAs
- epigenetic miRNAs
- biomarkers
- dysregulated expression
1. Introduction
Micro RNAs (miRNAs) are a subset of human non-coding RNA (ncRNA) that plays an essential role in regulating gene expression, RNA maturation, and protein synthesis [1][2][1,2]. ncRNAs have long been considered as “junk” elements; they account for about 75–90% of the human genome and are classified in two main groups according to their length: small (<200 nucleotides; miRNAs) and long (>200 nucleotides; lncRNAs) [2][3][2,3]. About 19–22 nt that play a crucial role in cell growth, development, and differentiation by regulating gene expression [4][5][4,5].
Most human miRNAs map in the introns of coding genes; some may overlap with the exons, less frequently in non-coding regions or next to the 3′-UTR sites, and they are often located in the co-transcribed clusters [6][7][8][6,7,8]. miRNA distribution in the human genome is not random, and some chromosomes such as 1, 2, 19, and X have higher numbers of miRNAs than others [9]. Several miRNAs are located next to fragile chromosomal sites or breakpoints frequently involved in leukemia rearrangements [10][11][10,11]. Most of the mature miRNAs bind to the “seed” region (5–8 nt long) in the 3′UTR of target mRNA molecules, induce silencing complex (miRISC) and act as post-transcriptional regulators causing mRNA degradation or translational repression via deadenylation, decapping, and exonucleolytic processes [12][13][14][15][14,15,16,17].
Moreover, other regions, such as gene promoters, 5′ UTR, or coding regions, may also be linked by miRNAs [16][17][18,19]. One miRNA can inhibit many different mRNA transcripts, often with similar functions, and control multiple signaling pathways [18][22]; conversely, one mRNA transcript can be targeted by several miRNAs [19][23]. miRNA expression is highly tissue-specific, some miRNAs being expressed in a specific cell or tissue type; deregulation of miRNA expression has been associated with several diseases and cancers. Almost 50% of miRNAs are located near or within genes translocated in cancer [20][24].
By down-regulating the expression of oncogenes or tumor suppressors, miRNAs can prevent or promote tumorigenesis; therefore, they may act as oncogenes (onco-miRs) or tumor suppressors [21][25]. Abnormal expression of miRNA has been reported in many malignancies, in which tumor suppressors are downregulated and oncogenic The roles of miRNA in leukemia pathogenesis are still emerging, and several studies have suggested miRNA expression profiles using as biomarkers for diagnosis, prognosis, and response to therapy in leukemia. These aspects will be discussed in the present review considering their mechanisms of action and the miRNAs most frequently deregulated in myeloid or lymphoid leukemias.
2. Epigenetic miRNAs (epimiRNAs)
However, some other miRNAs, defined as epigenetic-miRNAs (epi-miRNAs or epi-miRs), can directly or indirectly influence the expression of known epigenetic regulators such as DNA methyltransferases (DNMTs), HDACs, and components of PRC [22][23][29,30]. The first identified example of epi-miRNAs was the miR-29 family members (29a, 29b, and 29c); they regulate the expression of DNMT3A and DNMT3B in lung cancer and acute myeloid leukemia (AML). In vitro experiments showed that the exogenous introduction of these miRNAs in lung cancer or AML cell lines led to the reversion of the neoplastic phenotype by inhibiting different DNA methyltransferases and the consequent hypomethylation and reactivation of other target genes [24][25][26][31,32,33]. Increasing the expression of this epi-miRNA, apoptosis and differentiation are stimulated in leukemic cells due to inhibition of the AML1-ETO and other epigenetic regulators [27][37].
3. miRNAs and lncRNAs Interaction
Several data showed that many lncRNAs have multiple miRNA response elements (MRE), that are regions mediating reciprocal interaction; lncRNAs could therefore influence miRNA expression as they can act as sponges in both normal and cancer cells, determining several interactive networks. [28][67].HOTAIRM1also showed an essential role in the pathogenesis of AML cases with the t(15;17) translocation, acting as a microRNA sponge sequestering several miRNAs [29][68]. Several recent papers reported different miRNA and lncRNA interactions in AML, such as LINC01018 and miR-499a-5p, with LINC01018 acting as a sponge of miR-499a-5p, which in turn targets PDCD4 gene [30][69]. Both LINC01018 overexpression and miR-499a-5p knockdown suppressed AML cell proliferation and induced apoptosis, whereas miR-499a-5p transfection and silencing ofPDCD4reversed these effects [30][69].
Moreover, lncRNA and miRNA interaction seem to be frequently involved in chemoresistance mechanisms in AML, as for lncRNA-UCA1/miR-125a/hexokinase 2 orHOAX-AS2/miR-520c-3p [31][32][71,72]. UCA1 expression was found upregulated following adriamycin (ADR)-based chemotherapy, and UCA1 knockdown. On the other hand, lncRNAHOXA-AS2 was significantly upregulated in BM cells from AML cases after ADR-based chemotherapy, and its knockdown inhibited cell proliferation and induced apoptosis. These data showed that both lncRNAsUCA1 and HOAX-AS2 may represent useful therapeutic targets for overcoming ADR-chemoresistance in AML [32][72].
Another interesting example is represented by the network between lncRNAs and MYC expression in both myeloid and lymphoid malignancies, which contributes to inhibiting apoptosis, stimulates cell proliferation, induces genomic instability and resistance to therapy [33][73]. In AML, lncRNACCAT1 interacts and inhibits miR-155, whose targets are MYC, AP-1, FOS, and c-JUN, which regulate myeloid cell differentiation [34][35][74,75]. CCAT1 sponges miR-155 and stimulates MYC expression; interestingly, in previous studies, CCAT1 has been reported to be activated by MYC, suggesting the existence of aMYC/CCAT1/miR-155 feedback loop [35][75]. Other examples of lncRNA and miRNA interaction include MEG3/miR-147 andUCA1/mir-16 that have been detected in CML and are considered possible therapeutic targets in blast crisis or imatinib (IM) resistance, respectively [36][37][76,77]. Moreover, in lymphoma, the interaction between HOTAIR and miR-148b regulates apoptosis and the cell cycle progression of B cells.
4. miRNAs Expression Profile in Leukemia
as different miRNAs are expressed in a specific cell or tissue type; miRNAs play a crucial role in regulating gene expression during normal hematopoiesis, acting on the self-renewal capacity of hematopoietic stem cell (HSC) and the differentiation of lineage-restricted progenitors [38][39][91,92]. Thirty-three miRNAs were identified as specifically expressed in CD34+ hematopoietic stem-progenitor cells (HSPCs) [40][93]. Some miRNAs such as miRNA-17, -24, 146, -155, -128, and -181, were found to be expressed in early hematopoietic cells whereas other miRNAs, such as miRNA155, were able to control specific processes, as myelopoiesis and erythropoiesis [40][93], or as miR-34a and miR-17-92 clusters that have an essential role in the pro- to pre-B-cell differentiation [41][94]. miR-181 cluster also plays a critical role in the differentiation of hematopoietic cells as T, B, and natural killer cells or megakaryocytes [42][95].
Deregulation of miRNA expression has been associated with several diseases and cancers, and specific miRNA expression profiles have been reported in several hematologic neoplasms [43][44][96,97]. Microarray technologies, next-generation sequencing (NGS), and quantitative real-time PCR (qRT-PCR) are the most valuable methodologies for identifying reproducible dysregulated expression profiles in specific leukemia types (Figure 1). Unlike microarray analysis, NGS by miRNA-Seq shows high sensitivity in discovering new miRNAs and detecting whole-genome miRNA transcripts (miRNoma) with no need for previous selection. Several of these miRNAs may potentially be used as prognostic biomarkers, either as single miRNAs or as miRNA expression profiles.
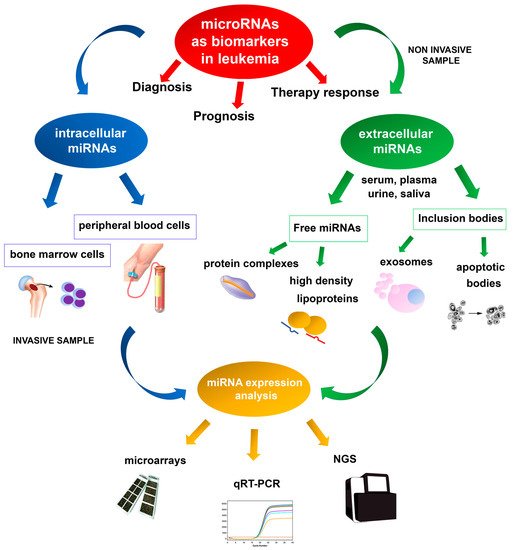
The first evidence of the involvement of miRNAs in leukemogenesis was reported in a study of CLL aimed at characterizing a 30 Kb deletion on 13q14; the study did not identify any protein-coding genes, but a cluster of two miRNAs, miR-15a and miR-16-1, was found to be deleted or down-regulated in most of the CLL cases examined [45][98]. Moreover, these miRNAs have also been shown to be frequently deregulated in other solid and hematological cancer types [46][47][48][99,100,101], and can modulate cell cycle progression and induce apoptosis by targeting pivotal genes such as BCL2, MCL1, CCND1, or WNT3A [49][50][51][102,103,104].
Different studies reported specific miRNA expression profiles that distinguish between AML and acute lymphoblastic leukemia (ALL), some miRNAs being reported in different studies, such as miR-23a, miR-27a/b, miR-128a, miR-128b, miR-221, miR-222, miR-223, and let-7b [52][53][105,106]. In a study by Mi et al., four miRNAs were sufficient to distinguish between AML and ALL with an accuracy of greater than 95%, as let-7b/miR-223 being significantly upregulated and miR-128a/miR-128b downregulated in AML comparing to ALL [52][105]. Together, the above work showed that these identified miRNAs could be new potential markers for ALL and AML classification and diagnosis [54][62].
One of the miRNAs most frequently involved in AML pathogenesis is miR-155, which is also commonly overexpressed in B-cell neoplasms, where it is considered an oncogenic driver of B-cell lymphoma [55][107]. This miRNA shows a contrasting dose-dependent function as onco-miRNA or tumor suppressor according to the expression levels. In short, a high level of expression is correlated with the antitumor effect and inhibition of AML cell proliferation. In contrast, an intermediate-low level of expression induces oncogenesis [56][108] and has been associated with poor prognosis in AML irrespective of specific cytogenetic or molecular aberrations [56][108].
Several studies have identified specific miRNA signatures in different AML subgroups defined by the classification of myeloid neoplasms [57][109] (Table 1), and a correlation was detected with cytogenetics alterations [58][59][60][110,111,112], prognosis, and clinical characteristics [61][65].
| AML | miRNAs | Expression Data | References |
|---|---|---|---|
| t(15;17)(q24;q21) PML-RARA | miR-127, miR-154, miR-154∗, miR-299, miR-323, miR-368, miR-370, miR-382, miR-134, miR-376a, miR-127, miR-299–5p, miR-323, miR-224 | Upregulation of miRNAs mapping in in the 14q32 imprinted region | [58][59][60][110,111,112] |
| t(8;21)(q22;q22.1) RUNX1-RUNX1T1 | miR-126, miR-146a, miR-133a, let-7b, let-7c | Overexpression of miR-126 and miR-146a; downregulation of miR-133a let-7b and let-7c | [59][60][62][111,112,113] |
| inv(16)(p13.1q22) CBFB-MYH11 | miR-99a, miR-100, miR-224, miR-126 | miRNA signature sometimes overlapping with t(8;21) AML | [58][59][60][110,111,112] |
| FLT3-ITD | miR-155, miR-10a, miR-10b | Upregulation | [59][63][111,114] |
| Mutated NPM1 | miR-10a and b, let-7, miR-29, miR-15a/16-1, miR-17-18a-19a-20a, miR-204 and miR-128a | Upregulation of miR-10a and b, let-7, miR-29, miR-15a/16-1, and miR-17-18a-19a-20a; downregulation of miR-204 and miR-128a | [63][64][65][114,115,116] |
| MLL rearranged | miR-34b, miR-15a, let-7 family, miR-196, miR-10a, miR-331, and miR-340, miR-17-92, miR-126, -126∗, -224, -368, -382, 17-5p, and -20a | Downregulation of miR-34b, miR-15a, let-7, and miR-196; upregulation of miR-17-92, miR-126, -126∗, -224, -368, -382, 17-5p, and -20a | [58][60][66][110,112,117] |
| Mutated IDH2 | miR-125b, miR-1, miR-133, miR-194-1, miR-526, miR-520a-3p, miR-548b | Upregulation of miR-125b, miR-1 and miR-133; downregulation of miR-194-1, miR-526, miR-520a-3p, and miR-548b | [67][118] |
| Mutated RUNX1 | miR-223, let-7, miR-211, miR-220, miR-595 | Downregulation of miR-223 and let-7; upregulation of miR-211, miR-220, and miR-595 | [68][119] |
| Normal karyotype | miR-181a/b, miR-124, miR-128-1, miR-194, miR-219-5p, miR-220a, miR-320 | Upregulation | [69][120] |
| ALL | miRNAs | Expression Data | References |
| MLL rearranged | miR-128b, miR-708, let-7b | Downregulation | [52][70][105,121] |
| t(12;21)(p13;q22) ETV6-RUNX1 | miR-100, miR-125b, miR-99a, miR-126, let-7c, miR-181a | Upregulation of miR-100, miR-125b, miR-99a, miR-126, let-7c; downregulation of miR-181a | [71][72][73][74][122,123,124,125] |
| t(9;22)(q34;q11) BCR-ABL1 | miR-125b-2, miR-203 | Overexpression of miR-125b-2; downregulation of miR-203 | [75][126] |
| hyperdiploid karyotype | miR-222, miR-223, miR-374, miR-660, miR-98 and miR-511 | Upregulation | [71][122] |
| t(1;19)(q23;p13) TCF3-PBX1 | miR-126, miR-146a, miR-511, miR-545, miR-365, miR-24, miR-30d, miR193, miR-181, miR-708 | Downregulation of miR-126, miR-146a, miR-511, miR-545, miR-365, miR-24, miR-30d, miR193; upregulation of miR-181, miR-708 | [72][76][123,127] |
| T-ALL | miR-17-92, miR-708, miR-196b, miR-128, miR-181, miR-29, miR-150, miR-99a and miR-708 | Overexpression of miR-17-92, miR-708, miR-196b, miR-128, miR-181; Downregulation of miR-29 | [77][78][79][128,129,130] |
In AML with the t(15;17) translocation, the upregulation of miRNAs located in the 14q32 imprinted domain (miR-127, miR-154, miR-154∗, miR-299, miR-323, miR-368, and miR-370) was reported in a first study [58][110], whereas a set of partially overlapping strongly upregulated microRNAs (miR-382, miR-134, miR-376a, miR-127, miR-299–5p, and miR-323) was described by Jongen-Lavrencic et al. [59][111]. In another study, the overexpression of miR-224, miR-368, and miR-382 was detected [60][112].
AML with t(8;21) showed high miR-126/126∗ [60][112] and miR-146a expression with decreased miR-133a [58][110]; other evidence showed a set of down-regulated miRNAs, including two members of a known tumor suppressor microRNA family, let-7b and let-7c [59][111], that was previously found to be involved in other cancers [62][113].
In AML with inv(16), a high level of miR-99a, miR-100, and miR-224 expression or of miR-126/126∗ was observed by different investigators [58][60][110,112]. Overall, AML with inv(16) showed a miRNA signature that sometimes overlapped with t(8;21) AML; this is not surprising as both these AML subtypes belong to the CBF group [59][111].
In AML with FLT3-internal tandem duplication (FLT3-ITD), miR-155, miR-10a, and miR-10b were found to be upregulated [59][63][111,114].
In AML with NPM1 mutations, a specific miRNA-based expression signature was revealed with upregulation of miR-10a and b, members of the let-7 and miR-29 families, miR-15a/16-1 and miR-17-18a-19a-20a clusters, and downregulation of miR-204 and miR-128a, predicted to target HOX genes known to be upregulated inNPM1mutated AML [63][64][114,115]. A further study was based on an integrative approach based on both microRNA and gene expression profiles. Several interesting microRNA-target mRNA interactions, such as IRF2-miR-20a, KIT-miR-20a, and MN1-miR-15a, were identified. This study also showed a deregulated expression of tumor suppressor microRNAs, such as miR-29a and miR-30c, that seem to be involved in sensitivity to therapy [65][116].
In AML with balanced 11q23 translocations and KMT2A (MLL1) rearrangements, the downregulation of several tumor suppressor miRNAs such as miR-34b, miR-15a, the let-7 family, and miR-196, targeting several known target genes such as CDK4 and CCNE2, BCL2, RAS, and HOX genes was reported; other evidence showed that AML with the MLL rearrangement were characterized by the loss of miR-10a, miR-331, and miR-340 expression [58][110]. Other authors revealed the overexpression of miRNAs from polycistronic cluster miR-17-92, and a minimal class predictor with only seven miRNAs (miR-126, -126∗, -224, -368, -382, 17-5p, and -20a) was identified [60][112]. Leukemic cells with higher expression levels of miR-17-92 showed arrested differentiation and increased proliferation in concomitance with reduced expression of p21, a downstream target of polycistronic miR-17-92 [66][117].
In AML with IDH2 mutations, a specific signature for R172 IDH2-AML was identified with the upregulation of miR-125b, which targets theTP53gene and inhibits myeloid differentiation, miR-1 and miR-133, involved in embryonic stem-cell differentiation, and downregulation of miR-194-1, miR-526, miR-520a-3p, and miR-548b, not previously associated with normal hematopoiesis or AML [67][118].
In AML with RUNX1 mutations, miR-223 and two members of the let-7 tumor suppressor family were downregulated, whereas three miRNAs with an unknown role in leukemogenesis, miR-211, miR-220, and miR-595, were found to be upregulated [68][119].
MiRNA expression analysis performed on normal karyotype AML (CN-AML) revealed a prognostic relevant miRNA signature, as the upregulation of miR-181a/b and miR-124, miR-128-1, miR-194, miR-219-5p, miR-220a, and miR-320 was associated with a low or increased risk of failure to achieve complete remission (CR), of relapse or death, respectively [69][120]. Some miR-181 putative targets were genes involved in innate immunity, encoding interleukins, caspases, and Toll-like receptors [69][120].
miRNAs deregulation is a common event in B- and T-cell malignancies in which they act as either oncomiRs or tumor suppressors [41][71][80][94,122,131]. ALL [71][122]; miR-155, that induces pre-B cells clonal expansion and is overexpressed in different pediatric ALL subtypes [55][107]; miR-128b, that allows differentiation with AML cases and is downregulated in ALL with theMLL-AF4translocation [52][70][105,121]. Moreover, miR-203 is silenced through epigenetic mechanisms [75][126]; it has been shown that by enhancing miR-203 expression,BCR-ABL1transcript level is reduced, cell proliferation is inhibited, and resistance to TKI can be overcome [81][133]. Regarding ALL with hyperdiploid karyotype, miR-222, miR-223, miR-374, miR-660, miR-98, and miR-511 were found upregulated, probably as a consequence of their mapping location in chromosomes X and 10 that are frequently present as extra copies [71][122].
miR-708 was found upregulated when comparing T-ALL with healthy individuals and downregulated when comparing T-ALL with different leukemia subtypes [71][122]; miR-708 downregulation is a poor prognostic factor of T-ALL, as it induces an increased expression of CD47 and promotes the evasion of leukemic cells from macrophage-mediated phagocytosis [82][136]. Low levels of miR-150 expression were associated with poor prognosis as correlating with relapse, high-risk and high WBCs at diagnosis; the association of miR-150 downregulation and poor prognosis was also revealed in other hematologic malignancies as AML, CLL, and different lymphoma subtypes [41][94]. Regarding miR-99, different studies revealed that upregulation and downregulation were correlated with favorable and poor prognosis, respectively [83][84][138,139]. Finally, miR-708 upregulation was associated with a good prognosis, as lower relapse risk, low WBC count, and better overall survival were detected in ALL cases; on the contrary, miR-708 downregulation was revealed in poor prognosis subtypes as T-ALL and cases with MLL gene rearrangement [71][84][122,139].
Moreover, recent data revealed that miR-300 is a tumor suppressor miRNA inducing quiescence in CML leukemic stem cells (LSCs) [85][141], and that miR-126-3p influences both quiescence and self-renewal of CML LSCs [86][142] (Table 2). Another recent study showed a global decrease in microRNA levels in LSC-enriched CD34+CD38−CD26+and HSC from CML-CP patients compared to those from healthy donors HSC [87][143]. Edurne San José-Enériz et al. identified a group of 19 miRNAs that may predict clinical resistance to IM in patients with newly diagnosed CML [88][151]. Another study revealed that miR-30 induces the degradation of BCR/ABL1 mRNAs by binding directly to their 3′UTR, which was downregulated in CML patients less responsive to IM [89][152].
| CML | miRNAs | Expression Data | References |
|---|---|---|---|
| pathogenesis | miR-155, miR-300, miR-126-3p | Upregulation of miR-155, miR-300; Downregulation of miR-126-3p | [90][85][86][140,141,142] |
| TKI resistance | miR-30, miR-26a, miR-29c, miR-130b, miR-146a, miR-142-5, miR-365a-3p, miR-153-3p, miR-185 | Downregulation | [89][91][92][93][152,153,154,156] |
| TKI discontinuation | miR-148b and miR-215 | Downregulation | [89][91]153[92],154[93],156[94][152,,157] |
| CLL | miRNAs | Expression Data | References |
| 13q deletion | miR-15a/miR-16-1 cluster, miR-34a/miR-34b/miR-34c cluster | Downregulation of miR-15 and miR-16; upregulation of miR-34 cluster | [45]98[95],158[96][,15997][,160] |
| Trisomy 12 | miR-181a | Upregulation | [96][159] |
| 17p deletion | miR-15a, miR-21, miR-34a, miR-155, and miR-181b | Upregulation of miR-21, miR-34a, miR-155, miR-181b | [98][161] |
| 11q deletion | miR-34b/miR-34c cluster | Downregulation | [97][160] |
In contrast, another recent investigation did not find any significant differences in miRNA expression patterns between TKI responder and non-responder patients [99][162]. Different studies analyzing miRNA expression profiles at CML diagnosis and in cases showing resistance considered only a small series of CML cases and produced contrasting results. More extensive analyses are therefore needed to verify whether the aforementioned miRNAs may be used to discriminate between the responders and non-responders among CML patients as well as the predictive biomarkers for TKI resistance. The miRNA involvement has also been investigated in CML to identify possible biomarkers for TKI discontinuation; two miRNAs, miR-148b and miR-215, showed downregulated expression in CML cases with successful IM discontinuation, suggesting that these miRNAs may contribute to immune surveillance in CML patients showing safe TKI discontinuation [94][100][157,163].
miRNAs are involved in the regulation of B lymphocyte development and can be altered in different B-cell malignancies. Other miRNAs frequently involved in CLL and other B-cell malignancies are miR-150, miR-155, and the miR-17-92 cluster; they regulate the expression of crucial transcription factors involved in normal or malignant B-cells development [101][102][103][50,166,167]. miR-150 is considered a lymphopoietic-specific miRNA, as its overexpression inhibits the pro-B to pre-B transition, probably by targeting forkhead box P1 (FOXP1) and GRB2-associated binding protein 1 (GAB1), an important transcription factor involved in B-cell differentiation [104][105][106][168,169,170]. It has been shown that the treatment with “BCR inhibitor” ibrutinib induces the downregulation of several miRNAs involved in B cell activation, as miR-22, miR-34a, miR-146b, and miR-181b, whereas the expression of several target genes including ARID1BATM, HDAC1, CYLD, FOXP1, IBTK, ARID2, PTEN, and SMAD4 is activated [107][176].
5. Conclusions
Increasing evidence shows that miRNAs have a role as potential biomarkers in leukemia, allowing a better subtype classification, prognostic stratification, and predicting the response to treatment. An ideal biomarker should have a specific expression in patients compared to normal controls, should allow early diagnosis and minimal residual disease monitoring during the follow-up, with a possible non-invasive, simple and accurate detection method. About this, miRNAs can be extracted and analyzed from peripheral blood or bone marrow cells of leukemia patients, while circulating miRNAs can be examined by non-invasive methods based on liquid biopsy analysis and usually show expression profiles overlapping with neoplastic cells.
