Exosomes are a subgroup of extracellular vesicles that can be secreted by virtually all types of cells, including cardiomyocytes, cardiac fibroblasts, endothelial cells, and stem and progenitor cells.
- cardiac exosomes
- coding and non-coding RNA species
- cardioprotection
1. Exosomes
The International Society for Extracellular Vesicles (ISEV) classifies EVs as “the generic term for particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate”. Unless authors can establish specific markers of subcellular origin that are reliable within their experimental system(s), ISEV urges to consider the use of operational terms for EVs [1]. In this review, when referring to “exosomes”, we refer to a subgroup of EVs ranging in size from 30–150 nm [50][2].
Exosomes were first described in 1977 under the name prostasomes [51][3], and were given the name exosome in 1987 [52][4]. Exosomes have been identified in all body fluids, including serum, plasma, amniotic fluid, saliva, breast milk, urine, and in cell culture media, and are secreted by all cells [53,54][5][6]. Initially, exosomes were believed to be a way for the cells to discard waste [52][4], but are now regarded as a well-regulated form of intercellular communication. Exosomes are generated from the invagination of endosomes resulting in the formation of multivesicular bodies (MVBs), which then are secreted through fusion with the cell membrane into the extracellular space. They function as biological vehicles, transferring information between donor and target cells [55,56,57][7][8][9]. The composition and cargo of the exosomes change depending on the cell type of origin and different cellular conditions or treatments. Furthermore, exosomes have been shown to carry proteins, lipids, coding and non-coding RNA, and DNA [58,59,60][10][11][12]. In addition, exosomes are enriched in lipids, such as cholesterol, phosphatidylserine, sphingomyelin, glycosphingolipids, and ceramide. These are conserved and essential for maintenance of exosome morphology, exosome biogenesis, and regulation of homeostasis in target cells [61][13]. The precise selection of exosomal cargo is not yet completely understood, although studies indicate that exosome formation and protein sorting can be managed by the endosomal sorting complexes required for transport pathway (ESCRT) [62,63][14][15] or lipid raft-mediated pathway [64][16]. ESCRT is comprised of four complexes; ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, with multiple associated proteins, e.g., Vacuolar protein sorting-associated protein 4, Tumor susceptibility gene 101 protein, and programmed cell death 6-interacting protein [65][17]. The lipid bilayer of the exosomes protects their cargo, allowing them to persevere in the extracellular environment. Once secreted, exosomes travel into the bodily fluids to enter neighboring or distant target cells. It has been shown that target cells internalize exosomes through a variety of methods, e.g., ligand receptor binding, endocytosis, or membrane fusion [66][18].
2. Exosomes Derived from Cardiac Cells
Recently, the cardiovascular field has had an increasing interest in exosomes due to their aforementioned cardioprotective possibilities. Studies have proven that exosomes can carry many proteins that are relevant for cardioprotection, e.g., phosphatase and tensin homolog (PTEN), annexins, epidermal growth factor receptor (EGFR), TNF-α, and NAD(P)H oxidase [67][19]. Although cardiac cells are not considered typical secretory cells, a number of in vitro experiments using rodents have indicated that cardiomyocytes secrete exosomes both in healthy and ischemic conditions, which mediate communication between healthy and damaged cells [68][20]. This was first proven in 2007, where exosomes were showed to be released both in physiological and hypoxic conditions [69][21]. Exosomes derived from cardiomyocytes have been shown to modulate cell proliferation, migration, differentiation, survival, and angiogenesis in response to ischemic incidences [70,71][22][23]. There is an increasing number of observations showing that cardiomyocyte-derived exosomes are enriched with inflammatory factors such as TNF-α and IL-6 [72,73][24][25]. Additionally, studies have demonstrated that cardiomyocyte-derived exosomes contain multiple heat shock proteins (Hsp20, Hsp60 and Hsp70) [69,74,75][21][26][27]. Furthermore, exosomes derived from cardiomyocytes have been found to carry functional glucose transporter proteins (GLUT1, GLUT4) and glycolytic enzymes (lactate dehydrogenase) [74][26]. An overview of proteins found in exosomes is presented in Figure 31. In addition to cardiomyocytes, both cardiac fibroblasts, endothelial cells, and Cardiosphere-Derived Cells (CDC) has been shown to secrete exosomes.
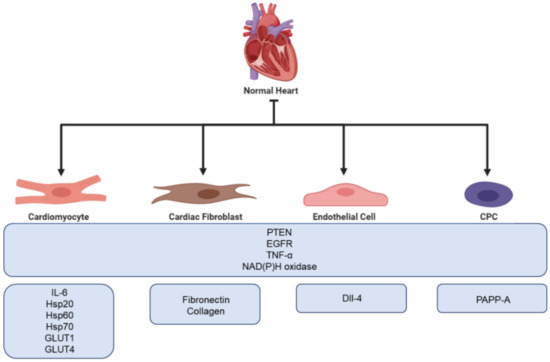
Figure 31. Overview of the cardioprotective proteins found in exosomes secreted from different cells of the myocardium.
A total of 60%–70% of all normal cardiac cells are made up of cardiac fibroblasts, which is one of the main building blocks of the extracellular matrix (ECM) [75][27]. During ischemia, cardiac fibroblasts become activated and involved in cardiac fibrosis and remodeling. Furthermore, with their secretory activity, they can influence the physiology of other cardiac cells [76][28]. Thus, fibroblasts play a crucial role in cardiac repair. However, excessive cardiac fibrosis is a major problem in most types of heart disease as it can interfere with normal heart function [77][29]. Proteomic analysis of cardiac fibroblasts from neonatal rats has revealed that cardiac fibroblast secreted exosomes are upregulated under hypoxic conditions. Furthermore, hypoxia has been proven to promote cardiac fibroblast exosomal enrichment in ECM proteins, e.g., fibronectin and collagen, as well as multiple mitochondrial associated proteins. This might indicate that cells use exosomes to modulate dysfunctional mitochondria during hypoxic stress [78][30]. Moreover, Abrila et al. [79][31] reported that the administration of cardiac fibroblast derived exosomes results in a 25% reduction in infarct size in a rat model of myocardial infarction compared to controls. Additionally, the presence of cardiac fibroblast derived exosomes in co-culture increased the viability of neonatal rat cardiomyocytes following hypoxia-reoxygenation injury. Furthermore, Hui et al. [80][32] have shown that cardiac fibroblast derived exosomes possess protective functions in cardiomyocytes both during acute myocardial infarction and ischemic post conditioning through the microRNA (miR)-423-3p/RAP2C pathway.
Endothelial cells form the endothelial barrier between blood and surrounding tissue and play a crucial role in the maintenance of cell homeostasis. When exposed to stress or damage, endothelial cells secrete cytokines, growth factors, and exosomes [81][33]. The cargo of endothelial exosomes has been shown to change under different conditions, e.g., hypoxia, often resulting in higher expression of proteins involved in ECM remodeling. Endothelial cells use exosomes for communication and to facilitate angiogenesis. Delta-like 4 factor (DII-4) has been identified in endothelial cell exosomes, promoting the increase in angiogenesis by inhibiting notch signaling. A study performed by Amabile et al. showed that surface biomarkers on endothelial exosomes correlate with cardiometabolic risk factors, e.g., CD144+ expression is significantly upregulated in patients suffering from hypertension [82][34]. Additionally, endothelial exosomes have been proven to play a role in the prevention of atherosclerosis through the Krüppel-like factor 2 (KLF2)-miR-143/145 pathway [83][35].
Cardiac-derived progenitor cells (CPCs) are a group of heterologous cells capable of responding to injuries and differentiating into new cardiac cells. If cultured in suspension, CPCs grow as spherical aggregates aptly named cardiospheres. It has been proven that both CPCs and cardiospheres are able to release exosomes possessing cardioprotective capacities. A recent study suggests that the presence of pregnancy-associated plasma protein-A (PAPP-A) on the surface of CPC exosomes aid in their cardioprotective capacity. PAPP-A functions by cleaving insulin-like growth factor binding protein-4 (IGFBP-4), which prompts the release of Insulin-like growth factor 1 (IGF-1), a key cardioprotective factor [84,85,86][36][37][38]. Furthermore, to investigate if exosomes released from CPC during hypoxia were able to protect the heart during in vivo IRI conditions, Gray et al. injected rats undergoing IR performed by ligation of the coronary with hypoxic derived CPC exosomes. They found that injection by CPC exosomes improved cardiac function by enhancing the tube formation of endothelial cells and decreased profibrotic gene expression resulting in delayed fibrosis [87][39].
3. The Reported Genomic Cargo of Cardiac Cell Derived Exosomes
Exosomes are also capable of transferring genetic materials, i.e., different types of RNA molecules, including microRNA (miR), messenger RNA (mRNAs), long non-coding RNA (lncRNA), and circular RNA (circRNA). Among the genetic materials detected in exosomes, miRs have received considerable attention in the context of cardiovascular disease. miRs are endogenous, short (17–25 nucleotides), and highly conserved non-coding RNA molecules, with key functions of fine-tuning gene expression by interfering with the translation of specific mRNAs at the post-transcriptional level [88][40]; hence miR are important components in the pathogenesis of heart failure as well as adaptive and maladaptive cardiac remodeling.
Recently, several specific circulating miRs have been associated with the development of HF. Reduced levels of circulating miRs, e.g., miR-18a, miR-27a, miR-30e, miR-26b, miR-199a, miR-106a, miR-652, let-7i, miR-18b, miR-18a, miR-223, miR-301a, miR-652, and miR-423 have been found in patients with HF, whereas the development of HF following ischemia has been associated with increased levels of miR-1, miR-133, miR-21, miR-29b, miR-192, miR-194, miR-34a, miR-208, miR-499, miR-423, miR-126, miR-134, miR-328, and miR-486, and decreases in miR-106, miR-197, and miR-223 (reviewed in [89][41]). Likewise, the levels of miR-144 have been found to increase by 1.6-fold in healthy human subjects undergoing a RIPC protocol [90][42]. In addition, a recent study performed on a large-scale study that included two independent cohorts of 2203 HF patients found that increased levels of miR-1254 and miR-1306 were associated with increased risk of death and hospitalization [91][43].
Studies on exosome secretion show that exosomes released from cells exposed to hypoxia are enriched with specific miRs, more specifically miR-126 and miR-210 [92][44]. Furthermore, a study performed by Matsumoto et al. on post myocardial infarction patients reported that serum levels of p53-responsive miRs (including miR-192, miR-195, and miR-34a) were significantly higher in the exosome fractions from HF patients than those of the control group [93][45]. Many studies have identified specific cardiomyocyte related genetic material within secreted exosomes. Wang et al. demonstrated high levels of miR-320 in exosomes secreted from cardiomyocytes of diabetic patients [94][46]. Another study has found that exosomes from cardiomyocytes are enriched with miR-29b, miR-323-5p, miR455, and miR-466 [95][47]. Additionally, miR-27a, miR28-3p, miR-34a, and miR-208a have been found to be highly expressed in cardiomyocytes and preferentially incorporated into exosomes [96,97][48][49]. It is worth mentioning that miR markers have also been identified in exosomes derived from other cells of the heart, e.g., cardiac fibroblast (miR-21* [98][50]) and endothelial cells (miR-143 and miR-145 [83][35]). An overview of miR in ischemic heart disease found is presented in Table 1.
Table 1.
MicroRNA (miR) in ischemic heart disease.
| miR ID | Change in Expression | Pathology | Reference |
|---|
| miR-18a | ↓ | HF | [89] | [41] |
| miR-27a | ↓ | HF | [89] | [41] |
| miR-30e | ↓ | HF | [89] | [41] |
| miR-26b | ↓ | HF | [89] | [41] |
| miR-199a | ↓ | HF | [89] | [41] |
| miR-106a | ↓ | HF | [89] | [41] |
| miR-652 | ↓ | HF | [89] | [41] |
| let-7i | ↓ | AHF | [99] | [51] |
| miR-18a | ↓ | AHF | [99] | [51] |
| miR-18b | ↓ | AHF | [99] | [51] |
| miR-223 | ↓ | AHF | [99] | [51] |
| ↓ | AMI | [89] | [41] | |
| miR-301a | ↓ | AHF | [99] | [51] |
| miR-652 | ↓ | AHF | [99] | [51] |
| miR-423 | ↓ |
lncRNAs are >200 nucleotides long non-coding transcripts, which are essential for the regulation of tissue homeostasis. The human genome has been found to contain over 50,000 lncRNAs, located within introns or antisense transcripts of coding genes, overlapping exons of coding genes or their promoters, or between genes [100][52]. Some lncRNAs have been shown to regulate the expression of coding genes at both the post-transcriptional and transcriptional level by directly binding to components of mRNAs and/or miRs. Other lncRNAs function as scaffolds for chromatin-modifying factors and thus regulate epigenetics [101][53]. Increasing evidence suggests that lncRNA plays important roles in cardiovascular diseases, e.g., lncRNA has been shown to be downregulated after acute myocardial infarction and upregulated during the later stages of HF. This indicates that lncRNAs are associated with post-infarction cardiac remodeling and chronic HF [102][54]. In a study, Greco et al. found that 13 lncRNAs were significantly modulated, 10 up- and 3 down-regulated, in HF patients compared to the control group. Greco et al. also found that the lncRNAs; Cyclin Dependent Kinase Inhibitor 2B antisense RNA 1 (CDKN2B-AS1), eosinophil granule ontogeny transcript (EGOT), H19 Imprinted Maternally Expressed Transcript (H19), HOX Transcript Antisense Intergenic RNA (HOTAIR), Limbic System Associated Membrane Protein antisense RNA 3 (LOC285194), RNA Component Of Mitochondrial RNA Processing Endoribonuclease (RMRP), Ro60-Associated Y5 (RNY5), SRY-Box Transcription Factor 2 overlapping Transcript (SOX2-OT), and Steroid Receptor RNA Activator 1 (SRA1) were significantly modulated in both end- and non-end-stage HF patients [103][55]. Furthermore, the concentration of Zinc Finger NFX1-Type Containing 1 Antisense RNA 1 (ZFAS1), which is known as a heart-specific lncRNA, has been found to be significantly reduced within patients with AMI compared with healthy volunteers and non-AMI patients [104][56]. ZFAS1 expression has also been found to be increased in the myocardium of AMI patients and in cultured neonatal mouse cardiomyocytes after exposure to hypoxia for 12 h. In addition, ZFAS1 has been shown to induce intracellular Ca2+ overload via alteration of Ca2+ transit in cardiomyocytes. It has also been shown that ZFAS1 has a strong affinity for sarcoplasmic reticulum Ca2+-ATPase 2a (SERCA2a), which is a key protein involved in the maintenance of normal intracellular Ca2+ [105][57]. Furthermore, HOTAIR, which is a modulator of HOX gene expression, has been shown by Goa et al. to be significantly decreased in the early phase of AMI compared with control groups. Similarly, the expression of HOTAIR in cardiomyocytes exposed to hypoxia for 1, 6, and 24 h was shown to be down-regulated [106][58]. Studies focused on lncRNA fibroblast growth factor 9-associated factor (FAF) show that FAF exerted significantly protective effects on cardiomyocytes exposed to hypoxia. In addition, FAF has been found to regulate the expression of Fibroblast Growth Factor 9 (FGF9), which is a known protective factor in post-MI. Thus, FAF can regulate apoptosis through positively controlling FGF9 by influencing the PI3K–AKT signaling pathway [105,107][57][59]. An overview of lncRNA in ischemic heart disease is presented in Table 2.
Table 2.
Long non-coding RNA (lncRNA) in ischemic heart disease.
| lncRNA | Pathology | Reference |
|---|
| CDKN2B-AS1 | HF | [103] | [55] | |||||
| EGOT | HF | [103] | [55] | |||||
| H19 | HF | [103] | [55] | |||||
| HOTAIR | HF | [103] | [55] | |||||
| LOC285194 | HF | [103] | [55] | |||||
| RMRP | HF | [103] | [55] | |||||
| RNY5 | HF | [103] | [55] | |||||
| SOX2-OT | HF | [103] | [55] | |||||
| SRA1 | HF | [103] | [55] | |||||
| AHF | ||||||||
| ZFAS1 | AMI | [ | ||||||
| [ | ||||||||
| 99 | ||||||||
| ] | [ | 51 | ] | |||||
| miR-21 | ↑ | SHF | [91] | [43] | ||||
| 104 | ] | [56] | ||||||
| HOTAIR | AMI | [106] | [58] | ↑ | AMI | [89] | [41] | |
| miR-1 | ↓ | SHF | [91] | [43] | ||||
| ↑ | AMI | [89] | [41] | |||||
| miR-1254 | ↑ | CHF | [91] | [43] | ||||
| miR-1306 | ↑ | CHF | [91] | [43] | ||||
| miR-133 | ↑ | AMI | [89] | [41] | ||||
| miR-29b | ↑ | AMI | [89] | [41] | ||||
| miR-192 | ↑ | AMI | [89] | [41] | ||||
| miR-194 | ↑ | AMI | [89] | [41] | ||||
| miR-34a | ↑ | AMI | [89] | [41] | ||||
| miR-208 | ↑ | AMI | [89] | [41] | ||||
| miR-499 | ↑ | AMI | [89] | [41] | ||||
| miR-423 | ↑ | AMI | [89] | [41] | ||||
| miR-126 | ↑ | AMI | [89] | [41] | ||||
| miR-134 | ↑ | AMI | [89] | [41] | ||||
| miR-328 | ↑ | AMI | [89] | [41] | ||||
| miR-486 | ↑ | AMI | [89] | [41] | ||||
| miR-106 | ↓ | AMI | [89] | [41] | ||||
| miR-197 | ↓ | AMI | [89] | [41] |
HF, heart failure; AHF, acute heart failure; AMI, acute myocardial infarction; SHF, systemic heart failure; CHF, chronic heart failure.
HF, heart failure; AMI, acute myocardial infarction.
circRNA, a type of lncRNA, consists of stable closed-ringed non-coding RNA molecules that are rich in miR-binding sites. As a result of this, circRNA can counteract the inhibitory effects on miRs on their target genes, thereby increasing the target gene expression levels. This is a mechanism known as the competitive endogenous RNA (ceRNA) mechanism. Previous findings reveal that some circRNAs are downregulated during myocardial infarction induced HF in mice, indicating a possible link between HF and circRNA [108,109][60][61]. A study focusing on the circRNA-expression profiles in peripheral blood samples from HF patients identified 56 differentially expressed circRNAs. Among the identified circRNA, hsa_circ_0097435 was found to be significantly more abundant in HF patients than in normal volunteers. Furthermore, hsa_circ_0097435 was shown to be encapsulated in exosomes. In the same study, it was demonstrated that hsa_circ_0097435 could promote apoptosis and associate with multiple miRs [110][62]. All these findings indicate that exosomes might have potential as biomarkers for cardiovascular disease.
4. Exosomes as Biomarkers
In acute pathologies, e.g., AMI, it is vital to rapidly identify and individualize the cardiac injury to optimize treatment strategies. Biomarkers are defined as measurable and quantifiable biological parameters that serve as indicators used for health and physiology assessments, such as disease risk and diagnosis [111][63]. A biomarker is considered good if it is easily measured and can be used as a surrogate marker for disease and its severity [112][64]. Ischemic related incidences are today commonly diagnosed using the cardiac biomarkers Cardiac Troponin T and I as well as Creatine-Kinase-MB [113][65]. Levels of Troponin I in blood have a peak 12 h after the ischemic damage to the heart and is proportional to the development of infarct size [111,114][63][66].
Shortly after the onset of injury, the heart can release characteristic exosomes of which their contents might be utilized for early diagnosis of cardiovascular diseases. Some have the capacity to replace already existing protocols, and others can be used in collaboration with classical analysis to help produce more accurate diagnoses. The discovery of exosomal miR has given rise to new possible biomarkers. An example is miR-208a, which, in a group of 66 patients (33 AMI patients and 33 non-AMI patients), was detectable in 0% of the healthy patients but detectable in 90.9% of all patients affected by AMI. The ROC curves of miR-208a between the AMI and non-AMI groups had an area under the curve (AUC) of 0.965. Furthermore, miR-208a was detected after 4 h from the onset of chest pain, which is very early compared to the appearance of detectable troponin [115][67]. Several other miRs have been shown to be upregulated in AMI patients, i.e., miR-208b, miR-1, miR-133a, and miR-499, but none were superior to the already existing troponin assays [116][68]. Moreover, in a study by Gidlöf et al., the circulating levels of miR-208b and miR-499-5p were assessed in non-MI (n = 88) and MI patients (n = 319), with an AUC of 0.82 and 0.79, respectively. In addition, the results showed that the plasma levels of miR-208b and miR-499-5p were upregulated in correspondence to the increase in the risk of death of HF, giving an indication of the prognosis, thus further solidifying miRs as possible biomarkers [117][69]. A study published by Matsumoto S et al. studying the expression of different miRs post-onset of AMI s (HF group, n = 21; control group, n = 65) suggests that exosome bound miRs can be used as predictive indicators of ischemic HF following AMI. Results showed that levels of p53-responsive miR-192, miR-194, and miR-34a were highly enriched in the exosome fraction in patients that developed HF post-AMI [93][45]. This is interesting as elevated p53 levels correlate with cardiomyocyte apoptosis and hypertrophy in end-stage human HF [118][70]. In addition to miRs, circRNAs have been shown to be regulated during cardiac development and failure, and may thus be suitable as potential biomarkers for HF. Moreover, the closed ring structure of circRNAs makes them more stable than linear RNA, further indicating their possibilities as useful biomarkers [119][71].
In addition to non-coding RNAs, several bioactive proteins such as Apolipoprotein D (APOD) and Apolipoprotein C3 (APOC3) (lipid metabolism), C1Q1A and C5 (complement activation), Glycoprotein Ib Platelet Subunit Alpha (GP1BA), and Pro-Platelet Basic Protein (PPBP) (platelet activation pathways) have been identified in exosomes from patients suffering from MI, and thus may be used as biomarkers for myocardial injury [120][72]. It was shown in a study by Yu X et al. that exosomes derived from cardiomyocytes situated to hypoxic conditions mediate TNF-α production [72][24]. Likewise, DeJong et al. reported that in vitro hypoxia and endothelial activation stimulate upregulation of certain proteins in exosomes, e.g., fibronectin, collagen, and lysyl-oxidase-like 2 (LOXL2) [121][73]. Other studies have demonstrated that exosomes derived from cardiomyocytes can contain increased levels of angiotensin II type 1 receptor (AT1R), which has been shown to play an important role in maintaining blood pressure and heart function [122][74]. Overview of cardioprotective factors presented in Figure 42.
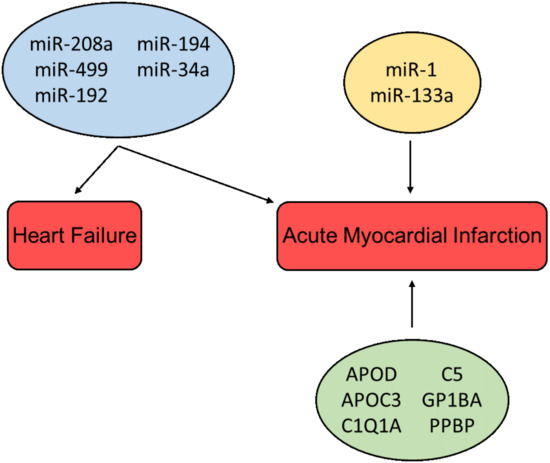
Figure 42. Cardioprotective factors associated with the diagnosis and prognosis of heart failure and acute myocardial infarction. miRs in the blue box are associated with multiple pathologies; miRs in the yellow box and proteins in the green box are associated with a single pathology.
The composition and biological contents of exosomes vary depending on the status of the mother cells at the time of exosome biogenesis. Thus, exosomes reflect the physiological status of their parent cells. Additionally, proteins and miRs carried by exosomes are more protected from degradation and the external environment than free-floating molecules in the blood. Therefore, exosomes may function as a more reliable source of biomolecules, preventing degradation and unstable detection results. Furthermore, by assessing the different expressions of surface proteins combined with the internalized cargo of exosomes, it is possible to better identify where the signals are coming from (Organ or cell specificity), resulting in a more specific diagnosis.
References
- Nowbar, A.N.; Gitto, M.; Howard, J.P.; Francis, D.P.; Al-Lamee, R. Mortality From Ischemic Heart Disease. Circ. Cardiovasc. Qual. Outcomes 2019, 12, e005375.
- Simons, M.; Raposo, G. Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581.
- Ronquist, G.; Hedström, M. Restoration of detergent-inactivated adenosine triphosphatase activity of human prostatic fluid with concanavalin A. Biochim. Biophys. Acta (BBA) Enzymol. 1977, 483, 483–486.
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420.
- Caradec, J.; Kharmate, G.; Hosseini-Beheshti, E.; Adomat, H.; Gleave, M.; Guns, E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin. Biochem. 2014, 47, 1286–1292.
- Slomka, A.; Urban, S.K.; Lukacs-Kornek, V.; Zekanowska, E.; Kornek, M. Large Extracellular Vesicles: Have We Found the Holy Grail of Inflammation? Front. Immunol. 2018, 9, 2723.
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208.
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500.
- Maisano, D.; Mimmi, S.; Russo, R.; Fioravanti, A.; Fiume, G.; Vecchio, E.; Nistico, N.; Quinto, I.; Iaccino, E. Uncovering the Exosomes Diversity: A Window of Opportunity for Tumor Progression Monitoring. Pharmaceuticals 2020, 13, 180.
- Choi, D.-S.; Kim, D.-K.; Kim, Y.-K.; Gho, Y.S. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom. Rev. 2015, 34, 474–490.
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120.
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079.
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857.
- Henne, W.M.; Stenmark, H.; Emr, S.D. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 2013, 5, a016766.
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565.
- Vidal, M.; Mangeat, P.; Hoekstra, D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J. Cell Sci. 1997, 110, 1867–1877.
- Juan, T.; Fürthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77.
- Dougherty, J.A.; Mergaye, M.; Kumar, N.; Chen, C.A.; Angelos, M.G.; Khan, M. Potential Role of Exosomes in Mending a Broken Heart: Nanoshuttles Propelling Future Clinical Therapeutics Forward. Stem Cells Int. 2017, 2017, 5785436.
- Yellon, D.M.; Davidson, S.M. Exosomes: Nanoparticles involved in cardioprotection? Circ. Res. 2014, 114, 325–332.
- Zhao, W.; Zheng, X.L.; Zhao, S.P. Exosome and its roles in cardiovascular diseases. Heart Fail. Rev. 2015, 20, 337–348.
- Gupta, S.; Knowlton, A.A. HSP60 trafficking in adult cardiac myocytes: Role of the exosomal pathway. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H3052–H3056.
- Vrijsen, K.R.; Sluijter, J.P.; Schuchardt, M.W.; van Balkom, B.W.; Noort, W.A.; Chamuleau, S.A.; Doevendans, P.A. Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. J. Cell. Mol. Med. 2010, 14, 1064–1070.
- Gennebäck, N.; Hellman, U.; Malm, L.; Larsson, G.; Ronquist, G.; Waldenström, A.; Mörner, S. Growth factor stimulation of cardiomyocytes induces changes in the transcriptional contents of secreted exosomes. J. Extracell Vesicles 2013, 2.
- Yu, X.; Deng, L.; Wang, D.; Li, N.; Chen, X.; Cheng, X.; Yuan, J.; Gao, X.; Liao, M.; Wang, M.; et al. Mechanism of TNF-α autocrine effects in hypoxic cardiomyocytes: Initiated by hypoxia inducible factor 1α, presented by exosomes. J. Mol. Cell. Cardiol. 2012, 53, 848–857.
- Datta, R.; Bansal, T.; Rana, S.; Datta, K.; Datta Chaudhuri, R.; Chawla-Sarkar, M.; Sarkar, S. Myocyte-Derived Hsp90 Modulates Collagen Upregulation via Biphasic Activation of STAT-3 in Fibroblasts during Cardiac Hypertrophy. Mol. Cell. Biol. 2017, 37.
- Garcia, N.A.; Moncayo-Arlandi, J.; Sepulveda, P.; Diez-Juan, A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc. Res. 2016, 109, 397–408.
- Xu, M.-Y.; Ye, Z.-S.; Song, X.-T.; Huang, R.-C. Differences in the cargos and functions of exosomes derived from six cardiac cell types: A systematic review. Stem. Cell Res. Ther. 2019, 10, 194.
- Bellin, G.; Gardin, C.; Ferroni, L.; Chachques, J.C.; Rogante, M.; Mitrečić, D.; Ferrari, R.; Zavan, B. Exosome in Cardiovascular Diseases: A Complex World Full of Hope. Cells 2019, 8, 166.
- Ranjan, P.; Kumari, R.; Verma, S.K. Cardiac Fibroblasts and Cardiac Fibrosis: Precise Role of Exosomes. Front. Cell Dev. Biol. 2019, 7, 318.
- Cosme, J.; Guo, H.; Hadipour-Lakmehsari, S.; Emili, A.; Gramolini, A.O. Hypoxia-Induced Changes in the Fibroblast Secretome, Exosome, and Whole-Cell Proteome Using Cultured, Cardiac-Derived Cells Isolated from Neonatal Mice. J. Proteome Res. 2017, 16, 2836–2847.
- Abrial, M.; Da Silva, C.C.; Pillot, B.; Augeul, L.; Ivanes, F.; Teixeira, G.; Cartier, R.; Angoulvant, D.; Ovize, M.; Ferrera, R. Cardiac fibroblasts protect cardiomyocytes against lethal ischemia-reperfusion injury. J. Mol. Cell. Cardiol. 2014, 68, 56–65.
- Luo, H.; Li, X.; Li, T.; Zhao, L.; He, J.; Zha, L.; Qi, Q.; Yu, Z. microRNA-423-3p exosomes derived from cardiac fibroblasts mediates the cardioprotective effects of ischaemic post-conditioning. Cardiovasc. Res. 2019, 115, 1189–1204.
- Sturtzel, C. Endothelial Cells. Adv. Exp. Med. Biol. 2017, 1003, 71–91.
- Amabile, N.; Cheng, S.; Renard, J.M.; Larson, M.G.; Ghorbani, A.; McCabe, E.; Griffin, G.; Guerin, C.; Ho, J.E.; Shaw, S.Y.; et al. Association of circulating endothelial microparticles with cardiometabolic risk factors in the Framingham Heart Study. Eur. Heart J. 2014, 35, 2972–2979.
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256.
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: Role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 2018, 114, 992–1005.
- Ashur, C.; Frishman, W.H. Cardiosphere-Derived Cells and Ischemic Heart Failure. Cardiol. Rev. 2018, 26, 8–21.
- Ibrahim, A.G.-E.; Cheng, K.; Marbán, E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014, 2, 606–619.
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263.
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355.
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084.
- Li, J.; Rohailla, S.; Gelber, N.; Rutka, J.; Sabah, N.; Gladstone, R.A.; Wei, C.; Hu, P.; Kharbanda, R.K.; Redington, A.N. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res. Cardiol. 2014, 109, 423.
- Bayés-Genis, A.; Lanfear, D.E.; de Ronde, M.W.J.; Lupón, J.; Leenders, J.J.; Liu, Z.; Zuithoff, N.P.A.; Eijkemans, M.J.C.; Zamora, E.; De Antonio, M.; et al. Prognostic value of circulating microRNAs on heart failure-related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur. J. Heart Fail. 2018, 20, 67–75.
- Varga, Z.V.; Zvara, Á.; Faragó, N.; Kocsis, G.F.; Pipicz, M.; Gáspár, R.; Bencsik, P.; Görbe, A.; Csonka, C.; Puskás, L.G. MicroRNAs associated with ischemia-reperfusion injury and cardioprotection by ischemic pre-and postconditioning: ProtectomiRs. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H216–H227.
- Matsumoto, S.; Sakata, Y.; Suna, S.; Nakatani, D.; Usami, M.; Hara, M.; Kitamura, T.; Hamasaki, T.; Nanto, S.; Kawahara, Y.; et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 2013, 113, 322–326.
- Wang, X.; Huang, W.; Liu, G.; Cai, W.; Millard, R.W.; Wang, Y.; Chang, J.; Peng, T.; Fan, G.C. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J. Mol. Cell. Cardiol. 2014, 74, 139–150.
- Chaturvedi, P.; Kalani, A.; Medina, I.; Familtseva, A.; Tyagi, S.C. Cardiosome mediated regulation of MMP9 in diabetic heart: Role of mir29b and mir455 in exercise. J. Cell. Mol. Med. 2015, 19, 2153–2161.
- Tian, C.; Gao, L.; Zimmerman, M.C.; Zucker, I.H. Myocardial infarction-induced microRNA-enriched exosomes contribute to cardiac Nrf2 dysregulation in chronic heart failure. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H928–H939.
- Yang, J.; Yu, X.; Xue, F.; Li, Y.; Liu, W.; Zhang, S. Exosomes derived from cardiomyocytes promote cardiac fibrosis via myocyte-fibroblast cross-talk. Am. J. Transl. Res. 2018, 10, 4350–4366.
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146.
- Ovchinnikova, E.S.; Schmitter, D.; Vegter, E.L.; Ter Maaten, J.M.; Valente, M.A.; Liu, L.C.; van der Harst, P.; Pinto, Y.M.; de Boer, R.A.; Meyer, S.; et al. Signature of circulating microRNAs in patients with acute heart failure. Eur. J. Heart Fail. 2016, 18, 414–423.
- Saddic, L.A.; Sigurdsson, M.I.; Chang, T.-W.; Mazaika, E.; Heydarpour, M.; Shernan, S.K.; Seidman, C.E.; Seidman, J.G.; Aranki, S.F.; Body, S.C.; et al. The Long Noncoding RNA Landscape of the Ischemic Human Left Ventricle. Circ. Cardiovasc. Genet. 2017, 10, e001534.
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59.
- Haemmig, S.; Simion, V.; Yang, D.; Deng, Y.; Feinberg, M.W. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr. Opin. Cardiol. 2017, 32.
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016, 14, 183.
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Li, K.; Liu, S.; Huang, Y.; Zhao, X.; Huang, L.; Wang, Z.; et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Sci. Rep. 2016, 6, 22384.
- Zhang, Y.; Jiao, L.; Sun, L.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca2+ Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ. Res. 2018, 122, 1354–1368.
- Gao, L.; Liu, Y.; Guo, S.; Yao, R.; Wu, L.; Xiao, L.; Wang, Z.; Liu, Y.; Zhang, Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell Physiol. Biochem. 2017, 44, 1497–1508.
- Shi, H.-J.; Wang, M.-W.; Sun, J.-T.; Wang, H.; Li, Y.-F.; Chen, B.-R.; Fan, Y.; Wang, S.-B.; Wang, Z.-M.; Wang, Q.-M.; et al. A novel long noncoding RNA FAF inhibits apoptosis via upregulating FGF9 through PI3K/AKT signaling pathway in ischemia–hypoxia cardiomyocytes. J. Cell. Physiol. 2019, 234, 21973–21987.
- Dou, Y.; Cha, D.J.; Franklin, J.L.; Higginbotham, J.N.; Jeppesen, D.K.; Weaver, A.M.; Prasad, N.; Levy, S.; Coffey, R.J.; Patton, J.G.; et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016, 6, 37982.
- Wu, H.J.; Zhang, C.Y.; Zhang, S.; Chang, M.; Wang, H.Y. Microarray Expression Profile of Circular RNAs in Heart Tissue of Mice with Myocardial Infarction-Induced Heart Failure. Cell Physiol. Biochem. 2016, 39, 205–216.
- Han, J.; Zhang, L.; Hu, L.; Yu, H.; Xu, F.; Yang, B.; Zhang, R.; Zhang, Y.; An, Y. Circular RNA-Expression Profiling Reveals a Potential Role of Hsa_circ_0097435 in Heart Failure via Sponging Multiple MicroRNAs. Front. Genet. 2020, 11, 212.
- Chan, D.; Ng, L.L. Biomarkers in acute myocardial infarction. BMC Med. 2010, 8, 34.
- Vasan, R.S. Biomarkers of Cardiovascular Disease. Circulation 2006, 113, 2335–2362.
- Li, L.-M.; Cai, W.-B.; Ye, Q.; Liu, J.-m.; Li, X.; Liao, X.-X. Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J. Emerg. Med. 2014, 5, 182.
- Antman, E.M.; Tanasijevic, M.J.; Thompson, B.; Schactman, M.; McCabe, C.H.; Cannon, C.P.; Fischer, G.A.; Fung, A.Y.; Thompson, C.; Wybenga, D.; et al. Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N. Engl. J. Med. 1996, 335, 1342–1349.
- Wang, G.-K.; Zhu, J.-Q.; Zhang, J.-T.; Li, Q.; Li, Y.; He, J.; Qin, Y.-W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666.
- Li, Y.Q.; Zhang, M.F.; Wen, H.Y.; Hu, C.L.; Liu, R.; Wei, H.Y.; Ai, C.M.; Wang, G.; Liao, X.X.; Li, X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 2013, 68, 75–80.
- Gidlof, O.; Smith, J.G.; Miyazu, K.; Gilje, P.; Spencer, A.; Blomquist, S.; Erlinge, D. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc. Disord. 2013, 13, 12.
- Mak, T.W.; Hauck, L.; Grothe, D.; Billia, F. p53 regulates the cardiac transcriptome. Proc. Natl. Acad. Sci. USA 2017, 114, 2331–2336.
- Altesha, M.A.; Ni, T.; Khan, A.; Liu, K.; Zheng, X. Circular RNA in cardiovascular disease. J. Cell Physiol. 2019, 234, 5588–5600.
- Cheow, E.S.; Cheng, W.C.; Lee, C.N.; de Kleijn, D.; Sorokin, V.; Sze, S.K. Plasma-derived Extracellular Vesicles Contain Predictive Biomarkers and Potential Therapeutic Targets for Myocardial Ischemic (MI) Injury. Mol. Cell Proteom. 2016, 15, 2628–2640.
- de Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell Vesicles 2012, 1.
- Pironti, G.; Strachan, R.T.; Abraham, D.; Mon-Wei Yu, S.; Chen, M.; Chen, W.; Hanada, K.; Mao, L.; Watson, L.J.; Rockman, H.A. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation 2015, 131, 2120–2130.
