Due to fast-developing molecular biology techniques, the identification of gene mutations is not laborious and expensive. HRM method is simple, fast, and costs less method to the identification of SNP compare with methods based on probes. In this topic, we will briefly describe the method, advantages, disadvantages, and limitations of HRM.
- HRM,
- SNP
- Genotyping
HRM (High-resolution melt) analysis is a powerful molecular biology detection method used for the identification of genetic mutations, DNA methylation analysis or species identification. HRM technic is based on PCR reaction with presence of fluorescence dye that binds to double-stranded DNA (dsDNA). The unbound dye shows a low level of fluorescence compared to dye bound to dsDNA when the fluorescence level increases greatly. After PCR reaction, the amplicon (usually 50 to 500 bp in length) is denatured gradually (this step is called melting analysis) from 65 °C to 95 °C with ramping by 0.2 °C. During this stage, the amplicon releases the fluorescent dye, allowing its detection. Amplicons from different samples with sequence changes (even differing by one base pair) will release fluorescent dye at different temperatures. A comparison with a reference control sample allows the sample to be assigned to the correct genotype group. The HRM method can discriminate DNA sequences based on their nucleotide composition, length, GC content, or strand complementarity and allows identification of deletions, insertions, and substitutions, as well as the detection of SNP (single nucleotide polymorphism) [1].
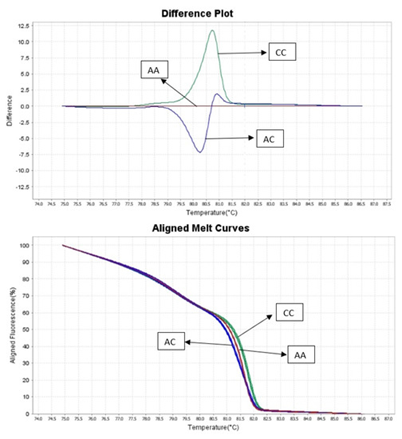
Fig. 1 HRM genotyping results.
Figure 1. HRM genotyping results.
HRM method as other genotyping method has various limitations described in the available literature [2,3,4][2][3][4]. One of the first and fundamental limitations affecting the sensitivity and reliability of the HRM method is DNA quality. In an experiment conducted by Smołucha et al. (2020), authors used DNA isolated from blood, which gives a good starting base for the quality and quantity of DNA obtained. Each sample was isolated using one method, normalized to the same concentration and diluted with the same buffer. This approach is consistent with that described by Słomka et al. [2] [2] in which the author analyzed the impact of DNA quality on genotyping efficiency by the HRM method. When using other methods such as TaqMan probes and Sanger sequencing an efficient DNA isolation method and standardization of its concentration is also recommended and can have an impact on the results.
Another important factor affecting HRM analysis is the correct design of primers. Primers should be prepared in accordance with accepted principles of primer design (a similar melting temperature for all primers, optimally 55–65 °C; avoiding secondary structures like hairpins, homodimers or heterodimers) [2]. It should also be remembered to normalize the concentration of primers before starting HRM analysis. Moreover, normalization procedure is also required using TaqMan probes as well as Sanger sequencing and influences the results. The cost of producing probes in the TaqMan method is much more expensive than the usual primers used in the HRM method, which ultimately impacts the total cost of conducting analyses. Features such as cost, speed, and quality of results are the basic criteria taken into account by breeders when choosing among available analyses.
Another important factor often discussed in the literature is the length of the amplicon. Products larger than 400 bp may have lower sensitivity than smaller amplicons. A sufficient amplicon length may be between 80 and 250 bp for routine analysis [1]. Technical problems and reproducibility of results are extremely important factors in any method [2]. In the HRM method, the reaction is carried out in one tube, which significantly reduces the possibility of contamination, as well as reduces the risk of errors by the operator. In the case of the Sanger sequencing reaction, the results obtained are a component of many stages (PCR reaction, enzymatic cleaning up of PCR product, sequencing reaction, capillary electrophoresis), which affects the final results in the form of a chromatogram.
- Vossen, R.H.; Aten, E.; Roos, A.; den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)—More than just sequence variant screening. Human Mutation2009, 30, 860–866
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. J. Mol. Sci.2017, 18, 316
- Tucker, E.J.; Huynh, B.L. Genotyping by High-Resolution Melting Analysis. In Crop Breeding; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; Volume 1145, pp. 59–66. ISBN 978-1-4939-0445-7.
- Montgomery, J.L.; Sanford, L.N.; Wittwer, C.T. High-Resolution DNA melting analysis in clinical research and diagnostics. Expert Rev. Mol. Diagn.2010, 10, 219–240.
References
- Vossen, R.H.; Aten, E.; Roos, A.; den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)—More than just sequence variant screening. Human Mutation2009, 30, 860–866
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. J. Mol. Sci.2017, 18, 316
- Tucker, E.J.; Huynh, B.L. Genotyping by High-Resolution Melting Analysis. In Crop Breeding; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; Volume 1145, pp. 59–66. ISBN 978-1-4939-0445-7.
- Montgomery, J.L.; Sanford, L.N.; Wittwer, C.T. High-Resolution DNA melting analysis in clinical research and diagnostics. Expert Rev. Mol. Diagn.2010, 10, 219–240.