Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Bruce Ren and Version 1 by Filippo Pesapane.
Radiomics is an emerging translational field of medicine based on the extraction of high-dimensional data from radiological images, with the purpose to reach reliable models to be applied into clinical practice for the purposes of diagnosis, prognosis and evaluation of disease response to treatment.
- radiomics
- breast cancer
- radiology
- oncology
- medical physics
- radiotherapy
- artificial intelligence
Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
In the last few years, the inclusion of standard digital imaging among the possible sources of big data for precision medicine has represented one of the new frontiers of research. Particularly, radiomics, the “omic” field related to diagnostic imaging, has been viewed as a great opportunity for several medical fields, yielding the most interesting results in oncology. Radiomic tumor analysis, including intra and inter-tumor heterogeneity, tumoral micro-environment and infiltrating cells, aims to extract quantitative features from medical imaging that are potentially beyond the perception of the human eye, in order to uncover novel features that are associated with treatment outcomes, disease molecular expressions or patient survival.
Moreover, recent technological advances in the field of computation and artificial intelligence (AI) applied to radiomics, hold promise in addressing challenges in its application [1,2][1][2]. In breast cancer, radiomics has been recently applied to identify molecular phenotypes and lymph node metastases, to evaluate treatment response and to predict disease survival [3,4,5,6,7,8][3][4][5][6][7][8].
2. Why Do We Need Radiomics in the Breast Cancer Care?
Breast cancer (BC) is the tumor with the highest incidence worldwide [9]. Although the screening and the advancements in personalized treatments have improved survival, it is estimated that BC related deaths will increase 43% globally from 2015 to 2030 [10].
At present, the diagnosis of early BC is based on radiological evaluation and histopathological confirmation of malignancy on biopsy samples [11,12][11][12]. With such approach is possible to characterize molecular alterations safely and effectively but have inherent limitations due to tumor heterogeneity and accessibility, as well as from procedure related risks [13,14][13][14].
Notably, the BC heterogeneity (which also undergoes temporal variation) is recognized as an important factor leading to cancer treatment failure and poor prognosis [15]. The quantification of heterogeneity relies on identification of various biomarkers, by the use of either tissue biopsy or medical imaging features. While the image-guided biopsies offer excellent spatial resolution for tissue analysis on a cellular scale and allow genetic and molecular sequencing [16]; however, biopsy is limited by risks of invasive procedures and focal sampling errors and limitation by tumoral characteristics such as small size, location or heterogenous necrosis [3,13,14][3][13][14].
Radiomics has the capability to analyze both temporal and spatial heterogeneities through quantitative serial data evaluation and recent advancements in radiomics analysis provide the potential to retrieve useful incremental information to characterize molecular alterations from standard imaging data in a non-invasive way [16]. However, radiomics currently suffers from lack of validation and standardization: further developments and improvements are needed to achieve reliable and clinically applicable results [14,16,17][14][16][17].
Whatever the method, the accurate biological assessment of BC is crucial because each subtype has its own biological and genetic profile with a subsequent different prognosis and treatment options. Subtypes are characterized by distinct molecular profiles, proliferation rates, tumor receptors and grade. Due to their potential effect on prognosis and clinical management, four biomarkers are tested consistently in biopsies and excision specimens of BC: estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and Ki67 antigen [11].
Prognosis for early-stage ER-positive/HER2-negative BC is usually excellent [18] and most of the invasive BCs hormone receptor positive show a more indolent clinical course [19]. Therefore, PR and ER status are considered as strong positive predictive factors since the advent of targeted hormone therapy.
At present, tumors are often classified as “luminal-A” or “luminal-B”, human epidermal growth factor receptor 2 (HER2)-overexpressing and triple negative (TN), based on immunohistochemical analyses [20,21][20][21]. Luminal A are cancers with ER+, PR+ and Ki67 < 20% and ER+, PgR+/− and Ki67 < 14%, and the best prognosis. Luminal B are cancers with ER+ but may have variable degrees of ER/PR expression, are higher grade and have higher proliferative fraction. HER2-overexpressing BC are ER−, PR− and HER2+ and they have a poorer prognosis than luminal BCs while the TN cancers (ER−, PR− and HER2−) have the poorest survival rate. These classifications could be used to inform adjuvant treatment decisions. Specifically, either grading or Ki-67 could be used to distinguish between the Luminal-A- and B-like [21].
Accordingly, biomarkers are crucial to tailor treatment strategies to the individual patient in the paradigm of personalized medicine [22]. However, the only way to obtain the biological profile of BC is currently through a tissue sample via surgery or biopsy. For this purpose, a new non-invasive technique based on imaging would be worthwhile. Radiomics, through the conversion of standard digital imaging into mineable, quantitative data expressing different tumor properties, has gained recognition as a new tool in the field of cancer care for non-invasively profiling of BC [1,3,7,8][1][3][7][8].
Particularly, the ultimate purpose of radiomics applied in BC care should be early diagnosis of BC and prediction of its clinical course and biological aggressiveness in order to optimize treatment [23].
The imaging evaluation of BC through mammography, ultrasound (US) or magnetic resonance imaging (MRI) is currently essentially qualitative. This includes subjective evaluations such as tumor morphology/structure, type of enhancement, anatomic relationship to the surrounding tissues. However, to reach a truly personalized medicine, a quantitative evaluation is demanded too [3]. Data derived from radiomics investigation, such as the intensity, shape, textural related features and wavelength related transforms, may provide valuable information to differentiate benign from malignant lesions, to predict treatment response, to assess cancer molecular profile and to derive robust models that combine multidisciplinary information [24,25,26,27,28,29,30,31][24][25][26][27][28][29][30][31].
3. The Workflow of a Radiomic Study
Most of radiomics studies concerns its application in the oncological field and the first step is generally to acquire the appropriate images. The Quantitative Imaging Biomarker Alliance and Quantitative Imaging Network have defined standardized imaging protocols and recommendations in the field of quantitative imaging [32] to improve the reproducibility of radiomics studies, which remains one of the biggest drawbacks currently limiting their clinical application.
Radiomics features are generally extracted from routine medical images that decode information about a region of interest (ROI) which are specified to limit the spatial extents of the analysis and can be delineated manually, semi-automatically or automatically, with increased reproducibility for textural features extracted with automatic segmentation algorithms compared to free-hand region delineation [33]. Feature extraction from the ROIs is performed using specific algorithms and are thus objective imaging features, with standard mathematical definition of the most common features [17].
An example of MRI-based radiomics workflow for features extraction is shown in Figure 1.
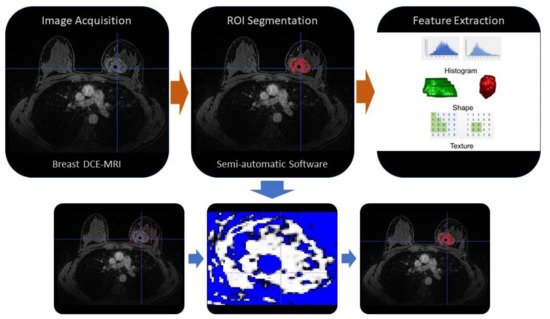
Figure 1. Example of MRI-based radiomics workflow. The first phase is the image acquisition (i.e., by breast MRI with contrast-enhancement sequences), then (orange arrow) the ROI segmentation could be performed manually or by automatic or semi-automatic software, finally (orange arrow) the radiomic features are extracted and selected by algorithms. An example of a semi-automatic segmentation by a threshold value method is shown in the three figures below (blue arrows). ROI: region of interest, DCE-MRI: Dynamic contrast enhancement-Magnetic resonance imaging.
The features can be broadly classified into four categories: morphological, histogram-based, textural and related to the gray level co-occurrence matrix and to transform-based features [32]. Morphological features describe different aspects of the lesion shape, such as volume, surface area, convexity or the borders heterogeneity. Histogram-based features characterize the histogram of voxel intensities, including the average value, standard deviation and parameters related to the histogram shape such as skewness and kurtosis. Textural features focus on the spatial arrangement of voxel intensities, trying to capture different properties of their distribution in terms of heterogeneity, randomness, presence of clusters or privileged signal directions. All these features can be calculated from the images as they are, or after applying mathematical transforms, such as wavelet of Laplacian of Gaussian (LoG), resulting in the so-called transform-based features. While hundreds or thousands of features may be computed, only a selection of fewer (and more specific) features is required to compute a clinically useful radiomic signature. Features whose value is not stable when images are repeatedly acquired under the same experimental condition (referred to as unstable or not repeatable features) should be identified a priori, by means of phantom studies or, if feasible, test-retest acquisitions in the clinical setting and eliminated [34]. Usually, a big gap between the number of features extracted (p) within a study and the number of patients actually recruited (n) remains, leading commonly to p>>n, with the risk to build radiomic models with high predictive accuracy in the experimental dataset but with extremely poor generalizability of the results, due to precise modelling of dataset “noise” instead of the true biological behavior. To overcome this problem feature selection and dimension reduction is of utmost importance, and different approaches can be performed, including rigorous algorithms such as principal component analysis, LASSO or Boruta [32]. The desired response variable differs based on the study, and models are built using the selected features to suit specific aims. For classification problems (e.g., benign vs malignant lesions), various classifiers are used including support vector machine (SVM), random forest (RF) and XGBoost classifiers. To predict continuous variables, such as the expression of biological markers, various regression methods including linear regression, regularized linear regression and RF are commonly used. For prediction of survival, Cox regression models with or without LASSO approach are finally performed.
Most radiomics studies involve a mixture of biomedical imaging specific techniques related to signal processing and proper AI applications, a broad field of computational techniques which includes machine learning (ML) and deep learning (DL) algorithms, the latter being often “black-box” and self-learning neural networks, with less dependence on human input in the model building step [33]. Given the high number of features obtained within radiomics studies and the often-non-linear relationships involved, these techniques offer a better approach in clinical predictive modeling compared to traditional inferential statistic and if properly applied, can limit model overfitting. Since a number of radiomics studies focused on BC are limited to single-center data lacking external validation, cross-validation with a leave-one-out, k-fold approach or with bootstrapping can be adopted using splits of the data into training and validation sets [33].
However, the optimal method of validation remains external dataset independent validation, which is typically accomplished in multi-center studies. However, acquiring multi-center data is challenging, so the solution may be to leverage an open database such as the cancer genome atlas program (TGCA), to acquire the external validation data.
As previously elucidated, reproducibility and standardization of radiomics analysis is currently the biggest issue. This partly because of the intrinsic high number of different steps involved and partly because every one of each can be performed in several different ways. The retrospective nature of studies, the heterogeneity of software and the variability of the radiomics features that can be extracted in the different studies raise legitimate concerns regarding the potential lack of reproducibility in radiomics. It is good practice to acquire imaging data using standardized settings that should be well documented in published papers, in order to be accurately evaluated during peer-review and be available to different research teams working on the same field. Data obtained under such settings should be shared on public repositories in order to receive appropriate external validation.
4. Radiomics Application in Breast Cancer
Even though there are studies about radiomics based on mammography, digital breast tomosynthesis (DBT), US and even PET/CT, in BC imaging scenario the radiomics approaches have been investigated mainly with MRI and, in the very last few years, with the contrast enhancement spectral mammography (CESM). However, results of most studies have been derived from relatively pure study designs, with homogeneous patient populations where the MRI was sourced from specific scanner systems and a single field strength. This limits their wider applicability and generalizability at present.
Table 1 and Table 2 summarize in our opinion the most relevant original studies and reviews, respectively, on radiomics in breast imaging published in peer reviewed journals from 01/2018 to 01/2021. Results from other interesting studies are briefly discussed only in the text.
Table 1. Original studies on radiomics in breast imaging published in peer reviewed journals from 01/2018 to 01/2021, classified on modality/technique and ordered by newest first. Relevant papers were obtained with a scoping review approach, using the following set of keywords and the relative controlled vocabulary terms (Mesh/Emtree): (radiomic* OR textur*) AND (breast) AND (cancer* OR malign* OR neoplas* OR metast* OR tumor* OR tumour*). The same approach was conduct for Table 2, which includes only review papers. ALN: axillary lymph node, AUC: area under the curve, BC: breast cancer, CESM: contrast enhancement spectral mammography, DCE: dynamic contrast-enhanced, ML: machine learning, NACT: Neoadjuvant Chemotherapy, pCR: pathological complete response, SD: standard deviation, TNBC: triple-negative breast cancer, T1WI: T1 weighted imaging, T2WI: T2 weighted imaging, VOI: volume of interest.
| Modality/Technique | Author | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion |
|---|---|---|---|---|---|---|
| CESM | Lin et al., 2020 [35] | Identification of benign and malignant BC lesions <1 cm | Radiomics features extracted from low-energy and recombined images on CC position | 139 patients | The radiomics nomogram combined with Radiomic-score, BI-RADS category and age showed AUC of 0.940. | The radiomics nomogram incorporated with CESM-based radiomics features, BI-RADS category and age could identify benign and malignant BC <1 cm |
| CESM | Mao et al. 2020 [36] | Pre-operative prediction of ALN metastasis | LASSO logistic regression was established for feature selection and utilized to construct radiomics signature | 394 patients | ROC curves of 0.774, 0.767 and 0.79 in the training, internal validation and external validation sets, respectively. | Authors identified the cutoff score in the radiomics nomogram as −1.49, which corresponded to a total point of 49 that could diagnose ALN metastasis with a sensitivity of >95%. |
| MRI | Tan et al., 2020 [6] | Value of radiomics feature extracted on the fat-suppressed T2WI for preoperative predicting ALN metastasis in BC | 17 texture features, 5 first-order statistical features, patient age, tumor size, HER2 status and thrombus | |||
| PET/CT | ||||||
| Ou et al., 2020 | ||||||
| [ | ||||||
| 49 | ||||||
| ] | ||||||
| Differentiating breast carcinoma from breast lymphoma | ||||||
| Radiomic features extracted with a local software | ||||||
| 44 BCs | ||||||
| AUCs of 0.867 and 0.806 for PET radiomic and clinical model, AUCs of 0.891 and 0.759 for CT based radiomic model on training and validation data. | Models based on clinical, and radiomic features of 18 F-FDG PET/CT images could accurately discriminate BC from breast lymphoma. |
Table 2. Review studies on radiomics in breast imaging published in peer reviewed journals from 01/2018 to 01/2021, ordered by newest first.
| Reference | Modality/Techique | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reig et al., 2020 [50] | MRI | Review focused on machine learning techniques in breast MRI | Pre-processing, neural networks, deep learning, machine learning, segmentation, texture analysis | Breast malignant and benign pathology. | ||||||||
| 329 BCs | ||||||||||||
| Sensitivity, specificity, accuracy and are under the curve value of radiomics signature 65.22%, 81.08%, 75.00% and 0.819. | The MRI-based radiomics signature and nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. | |||||||||||
| Choudhery et al., 2020 [5] | Assessment of BC molecular subtype, pCR and Residual Cancer Burden in BC Patients Treated with NACT | Morphological and three-dimensional MRI textural features were computed, including unfiltered and filtered image data, with different spatial scaling factors | 259 BCs | Differences in minimum signal intensity and entropy among the tumor subtypes were significant. Sphericity in HER2+ tumors and entropy in luminal tumors were significantly associated with pCR. Multiple features demonstrated significant association with pathological complete response and residual cancer burden in TNBC with SD of intensity achieving the highest AUC for pCR in TNBC. | MRI radiomics features are associated with different molecular subtypes of breast cancer, pathological complete response and residual cancer burden. | |||||||
| Hao et al., 2020 [37] | Contralateral BI-RADS 4 lesion assessment | 1046 radiomic features | 178 BCs | DCE-T1WI and T2WI imaging features signatures yielded an AUC of 0.77, which was better than the AUC of each signature alone. | The MRI radiomics-based ML model based on T2WI and DCE-T1WI features provided complementary information in discriminating benign and malignant contralateral BI-RADS 4 lesions. | |||||||
| Lo Gullo et al., 2020 [38] | Assessment of sub-centimetric breast masses in BRCA patients | Radiomics features calculated using open-source CERR software | 96 BRCA carrier | The ML model combining 5 parameters including clinical factors, GLCM-based correlation from the pre-DCE phases and first-order coefficient of variation from the 1st post-DCE phase, achieved a diagnostic accuracy of 81.5%. | Radiomics analysis improved diagnostic accuracy compared with qualitative morphological assessment alone. | |||||||
| Demircioglu et al., 2020 [39] | Molecular subtype, hormonal receptor status, Ki67- and HER2-expression, metastasis of lymph nodes and lymph vessel involvement as well as grading | 13.118 radiomic features extracted with a VOI-based approach | 98 BCs | PR and ER status predictions yielded AUCs of 0.67–0.69, Ki67 0.81 and HER2 Expressions 0.62. Involvement of the ALN could be predicted with an AUC of 0.80, while lymph node metastasis yielded an AUC of 0.71. | A rapid approach to VOI-based tumor-annotations for radiomics provides consisternt results to other studies in the same field. | |||||||
| The Author discuss the possible future directions of machine learning in the current workflow of breast lesions assessed with MRI. | ||||||||||||
| Granzier et al., 2019 [51] | MRI | Systematic review, response prediction of neoadjuvant therapy | Various radiomic feature models, evaluated with the Radiomics Quality Score (RQS) | Studies ranging between 35-414 BC | AUC values ranged from 0.83 to 0.85. The best performing multivariate prediction model, based on logistic regression analysis, showed AUC of 0.94. | The systematic review revealed large heterogeneity for each step of the MRI-based radiomics workflow. Consequently, the results are difficult to compare. | Zhang et al., 2020 [40] | Differentiation between benign and malignant lesions | Radiomics features extracted from T2WI, T1WI, DKI, ADC maps and DCE pharmacokinetic parameter maps | 207 BCs | The AUC of the optimal radiomics model, including T2 WI, DKI and quantitative DCE-MRI parameter maps was 0.921, with an accuracy of 0.833. | The model based on radiomics features from T2WI, DKI and quantitative DCE parameter maps has a high discriminatory ability for benign and malignant BC lesions. |
| Zhou et al., 2020 [41] | Differentiation between benign and malignant BC lesions | 99 texture and histogram parameters | 133 patients | The highest accuracy of 91% was achieved when using the smallest bounding box of peritumoral tissues in segmentation. | Using the smallest bounding box containing proximal peritumor tissue as input had higher accuracy compared to using tumor alone or larger boxes. | |||||||
| Liu et al., 2019 [42] | Assess lymphovascular invasion status | Radiomic signature composed of two features | 149 BCs | The value of AUC for a model combining both radiomic signature and ALN status (0.763) was higher than that for MRI ALN status alone and similar to that for the radiomics signature. | The DCE-MRI-based radiomics signature in combination with ALN status was effective in predicting the lymph and vascular invasion status of patients with BC before surgery. | |||||||
| Xie et al., 2019 [43] | Subtype classification of breast cancer | 2498 features extracted from the DCE and DWI, together with DCE images, changing over 6 time points and DWI images changing over 3 b-values | 134 invasive ductal carcinoma | Highest accuracy of 91% for comparing triple negative to non-triple negative cancers. | Whole-tumor radiomics on MRI provides a non-invasive approach for BC subtype classification. | |||||||
| Liang et al., 2018 [44] | Preoperative Ki-67 status | Radiomic features based on T2W and DCE-T1WI | 318 BC | The T2W image-based radiomics classifier showed significant discrimination for Ki-67 status, with AUC of 0.74 in the validation dataset. | The T2WI-based radiomics classifier was a significant predictor of Ki-67 status in patients with breast cancer while DCE-T1WI radiomic features were not able to discriminate Ki-67 status in the validation dataset. | |||||||
| Digital mammography | Tan et al., 2020 [45] | Pre-operative prediction of ALN metastasis | Radiomic signature nomogram combined with receptor status and molecular subtype | 216 BCs | The radiomics nomogram, comprising PR status, molecular subtype and radiomics signature, showed excellent calibration and better performance for the metastatic ALN detection (AUC 0.883 and 0.863 in the primary and validation cohorts), better than each independent clinical feature and radiomics signature. | The mammography-based radiomics nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. | ||||||
| Digital Mammography | Stelzer et al., 2020 [46] | Distinguish malignant from benign classification | 249 image features from gray-value histogram, co-occurrence and run-length matrices | 226 patients | A high sensitivity threshold criterion was identified in the training dataset and successfully applied to the testing dataset, demonstrating the potential to avoid 37.1-45.7 % of unnecessary biopsies at the cost of one false-negative. | Combined texture analysis and ML could be used for risk stratification in suspicious mammographic calcifications. | ||||||
| Zhou et al., 2019 [47] | HER-2 status | 186 radiomic features | 306 l BCs | In the testing set the AUC of the radiomic model in assessing HER-2 status was 0.787. | Radiomics features could help in the preoperative evaluation of HER-2 status in patients with BC. | |||||||
| Lei et al., 2019 [48] | Prediction of benign BI-RADS 4 calcifications | 8286 radiomic features extracted from the craniocaudal and mediolateral oblique scans | 212 calcifications | Six radiomic features and the menopausal state included in a radiomic nomogram could discriminate benign from malignant calcifications with an AUC of 0.80 in the validation cohort. | The mammography-based radiomic nomogram is a potential tool to distinguish benign calcifications from malignant calcifications. | |||||||
References
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35.
- Becker, A.S.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017, 52, 434–440.
- Pesapane, F.; Suter, M.B.; Rotili, A.; Penco, S.; Nigro, O.; Cremonesi, M.; Bellomi, M.; Jereczek-Fossa, B.A.; Pinotti, G.; Cassano, E. Will traditional biopsy be substituted by radiomics and liquid biopsy for breast cancer diagnosis and characterisation? Med. Oncol. 2020, 37, 29.
- Zhuang, X.; Chen, C.; Liu, Z.; Zhang, L.; Zhou, X.; Cheng, M.; Ji, F.; Zhu, T.; Lei, C.; Zhang, J.; et al. Multiparametric MRI-based radiomics analysis for the prediction of breast tumor regression patterns after neoadjuvant chemotherapy. Transl. Oncol. 2020, 13, 100831.
- Choudhery, S.; Gomez-Cardona, D.; Favazza, C.P.; Hoskin, T.L.; Haddad, T.C.; Goetz, M.P.; Boughey, J.C. MRI Radiomics for Assessment of Molecular Subtype, Pathological Complete Response, and Residual Cancer Burden in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Acad. Radiol. 2020, 20, 30607-3.
- Tan, H.; Gan, F.; Wu, Y.; Zhou, J.; Tian, J.; Lin, Y.; Wang, M. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Carcinoma Using Radiomics Features Based on the Fat-Suppressed T2 Sequence. Acad. Radiol. 2020, 27, 1217–1225.
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2019, 49, 74–80.
- Pinker, K.; Chin, J.; Melsaether, A.N.; Morris, E.A.; Moy, L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology 2018, 287, 732–747.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021.
- Tan, W.; Yang, M.; Yang, H.; Zhou, F.; Shen, W. Predicting the response to neoadjuvant therapy for early-stage breast cancer: Tumor-, blood-, and imaging-related biomarkers. Cancer Manag. Res. 2018, 10, 4333–4347.
- Tirada, N.; Aujero, M.; Khorjekar, G.; Richards, S.; Chopra, J.; Dromi, S.; Ioffe, O. Breast Cancer Tissue Markers, Genomic Profiling, and Other Prognostic Factors: A Primer for Radiologists. Radiographics 2018, 38, 1902–1920.
- Gradishar, W.J.; Anderson, B.O.; Balassanian, R.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; et al. Breast Cancer, Version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 310–320.
- Prud’homme, C.; Deschamps, F.; Allorant, A.; Massard, C.; Hollebecque, A.; Yevich, S.; Ngo-Camus, M.; Gravel, G.; Nicotra, C.; Michiels, S.; et al. Image-guided tumour biopsies in a prospective molecular triage study (MOSCATO-01): What are the real risks? Eur. J. Cancer 2018, 103, 108–119.
- Dercle, L.; Ammari, S.; Bateson, M.; Durand, P.B.; Haspinger, E.; Massard, C.; Jaudet, C.; Varga, A.; Deutsch, E.; Soria, J.C.; et al. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci. Rep. 2017, 7, 7952.
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345.
- Tselikas, L.; Sun, R.; Ammari, S.; Dercle, L.; Yevich, S.; Hollebecque, A.; Ngo-Camus, M.; Nicotra, C.; Deutsch, E.; Deschamps, F.; et al. Role of image-guided biopsy and radiomics in the age of precision medicine. Chin. Clin. Oncol. 2019, 8, 57.
- Zwanenburg, A.; Vallieres, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338.
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13.
- Bundred, N.J. Prognostic and predictive factors in breast cancer. Cancer Treat. Rev. 2001, 27, 137–142.
- Curigliano, G.; Burstein, H.J.; Winer, E.P.; Gnant, M.; Dubsky, P.; Loibl, S.; Colleoni, M.; Regan, M.M.; Piccart-Gebhart, M.; Senn, H.J.; et al. De-escalating and escalating treatments for early-stage breast cancer: The St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. 2019, 30, 1181.
- Viale, G.; Hanlon Newell, A.E.; Walker, E.; Harlow, G.; Bai, I.; Russo, L.; Dell’Orto, P.; Maisonneuve, P. Ki-67 (30-9) scoring and differentiation of Luminal A- and Luminal B-like breast cancer subtypes. Breast Cancer Res. Treat. 2019, 178, 451–458.
- Cain, E.H.; Saha, A.; Harowicz, M.R.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res. Treat. 2018, 173, 455–463.
- Rotili, A.; Trimboli, R.M.; Penco, S.; Pesapane, F.; Tantrige, P.; Cassano, E.; Sardanelli, F. Double reading of diffusion-weighted magnetic resonance imaging for breast cancer detection. Breast Cancer Res. Treat. 2020, 180, 111–120.
- Davnall, F.; Yip, C.S.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging 2012, 3, 573–589.
- Yip, S.S.F.; Parmar, C.; Kim, J.; Huynh, E.; Mak, R.H.; Aerts, H. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017, 97, 8–15.
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006.
- Rahmim, A.; Salimpour, Y.; Jain, S.; Blinder, S.A.; Klyuzhin, I.S.; Smith, G.S.; Mari, Z.; Sossi, V. Application of texture analysis to DAT SPECT imaging: Relationship to clinical assessments. Neuroimage Clin. 2016, 12, e1–e9.
- Pesapane, F.; Patella, F.; Fumarola, E.M.; Panella, S.; Ierardi, A.M.; Pompili, G.G.; Franceschelli, G.; Angileri, S.A.; Magenta Biasina, A.; Carrafiello, G. Intravoxel Incoherent Motion (IVIM) Diffusion Weighted Imaging (DWI) in the Periferic Prostate Cancer Detection and Stratification. Med. Oncol. 2017, 34, 35.
- Patella, F.; Franceschelli, G.; Petrillo, M.; Sansone, M.; Fusco, R.; Pesapane, F.; Pompili, G.; Ierardi, A.M.; Saibene, A.M.; Moneghini, L.; et al. A multiparametric analysis combining DCE-MRI- and IVIM-derived parameters to improve differentiation of parotid tumors: A pilot study. Future Oncol. 2018, 14, 2893–2903.
- King, A.D.; Chow, K.K.; Yu, K.H.; Mo, F.K.; Yeung, D.K.; Yuan, J.; Bhatia, K.S.; Vlantis, A.C.; Ahuja, A.T. Head and neck squamous cell carcinoma: Diagnostic performance of diffusion-weighted MR imaging for the prediction of treatment response. Radiology 2013, 266, 531–538.
- Peng, S.L.; Chen, C.F.; Liu, H.L.; Lui, C.C.; Huang, Y.J.; Lee, T.H.; Chang, C.C.; Wang, F.N. Analysis of parametric histogram from dynamic contrast-enhanced MRI: Application in evaluating brain tumor response to radiotherapy. NMR Biomed. 2013, 26, 443–450.
- Aerts, H.J. The Potential of Radiomic-Based Phenotyping in Precision Medicine: A Review. JAMA Oncol. 2016, 2, 1636–1642.
- Lee, S.H.; Park, H.; Ko, E.S. Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review. Korean J. Radiol. 2020, 21, 779–792.
- Bianchini, L.; Santinha, J.; Loucao, N.; Figueiredo, M.; Botta, F.; Origgi, D.; Cremonesi, M.; Cassano, E.; Papanikolaou, N.; Lascialfari, A. A multicenter study on radiomic features from T2-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics. Magn. Reson. Med. 2020, 85, 1713–1726.
- Mao, N.; Yin, P.; Li, Q.; Wang, Q.; Liu, M.; Ma, H.; Dong, J.; Che, K.; Wang, Z.; Duan, S.; et al. Radiomics nomogram of contrast-enhanced spectral mammography for prediction of axillary lymph node metastasis in breast cancer: A multicenter study. Eur. Radiol. 2020, 30, 6732–6739.
- Hao, W.; Gong, J.; Wang, S.; Zhu, H.; Zhao, B.; Peng, W. Application of MRI Radiomics-Based Machine Learning Model to Improve Contralateral BI-RADS 4 Lesion Assessment. Front. Oncol. 2020, 10, 531476.
- Lo Gullo, R.; Daimiel, I.; Rossi Saccarelli, C.; Bitencourt, A.; Gibbs, P.; Fox, M.J.; Thakur, S.B.; Martinez, D.F.; Jochelson, M.S.; Morris, E.A.; et al. Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers. Eur. Radiol. 2020, 30, 6721–6731.
- Demircioglu, A.; Grueneisen, J.; Ingenwerth, M.; Hoffmann, O.; Pinker-Domenig, K.; Morris, E.; Haubold, J.; Forsting, M.; Nensa, F.; Umutlu, L. A rapid volume of interest-based approach of radiomics analysis of breast MRI for tumor decoding and phenotyping of breast cancer. PLoS ONE 2020, 15, e0234871.
- Zhang, Q.; Peng, Y.; Liu, W.; Bai, J.; Zheng, J.; Yang, X.; Zhou, L. Radiomics Based on Multimodal MRI for the Differential Diagnosis of Benign and Malignant Breast Lesions. J. Magn. Reson. Imaging 2020, 52, 596–607.
- Liu, Z.; Feng, B.; Li, C.; Chen, Y.; Chen, Q.; Li, X.; Guan, J.; Chen, X.; Cui, E.; Li, R.; et al. Preoperative prediction of lymphovascular invasion in invasive breast cancer with dynamic contrast-enhanced-MRI-based radiomics. J. Magn. Reson. Imaging 2019, 50, 847–857.
- Xie, T.; Wang, Z.; Zhao, Q.; Bai, Q.; Zhou, X.; Gu, Y.; Peng, W.; Wang, H. Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Front. Oncol. 2019, 9, 505.
- Liang, C.; Cheng, Z.; Huang, Y.; He, L.; Chen, X.; Ma, Z.; Huang, X.; Liang, C.; Liu, Z. An MRI-based Radiomics Classifier for Preoperative Prediction of Ki-67 Status in Breast Cancer. Acad. Radiol. 2018, 25, 1111–1117.
- Tan, H.; Wu, Y.; Bao, F.; Zhou, J.; Wan, J.; Tian, J.; Lin, Y.; Wang, M. Mammography-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in breast cancer. Br. J. Radiol. 2020, 93, 20191019.
- Stelzer, P.D.; Steding, O.; Raudner, M.W.; Euller, G.; Clauser, P.; Baltzer, P.A.T. Combined texture analysis and machine learning in suspicious calcifications detected by mammography: Potential to avoid unnecessary stereotactical biopsies. Eur. J. Radiol. 2020, 132, 109309.
- Lin, F.; Wang, Z.; Zhang, K.; Yang, P.; Ma, H.; Shi, Y.; Liu, M.; Wang, Q.; Cui, J.; Mao, N.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for Identifying Benign and Malignant Breast Lesions of Sub-1 cm. Front. Oncol. 2020, 10, 573630.
- Zhou, J.; Tan, H.; Bai, Y.; Li, J.; Lu, Q.; Chen, R.; Zhang, M.; Feng, Q.; Wang, M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019, 121, 108718.
- Lei, C.; Wei, W.; Liu, Z.; Xiong, Q.; Yang, C.; Yang, M.; Zhang, L.; Zhu, T.; Zhuang, X.; Liu, C.; et al. Mammography-based radiomic analysis for predicting benign BI-RADS category 4 calcifications. Eur. J. Radiol. 2019, 121, 108711.
- Ou, X.; Zhang, J.; Wang, J.; Pang, F.; Wang, Y.; Wei, X.; Ma, X. Radiomics based on (18) F-FDG PET/CT could differentiate breast carcinoma from breast lymphoma using machine-learning approach: A preliminary study. Cancer Med. 2019.
- Zhou, J.; Zhang, Y.; Chang, K.T.; Lee, K.E.; Wang, O.; Li, J.; Lin, Y.; Pan, Z.; Chang, P.; Chow, D.; et al. Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J. Magn. Reson. Imaging 2019, 51, 798–809.
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine learning in breast MRI. J. Magn. Reson. Imaging 2020, 52, 998–1018.
- Granzier, R.W.Y.; van Nijnatten, T.J.A.; Woodruff, H.C.; Smidt, M.L.; Lobbes, M.B.I. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: A systematic review. Eur. J. Radiol. 2019, 121, 108736.
More
