Long noncoding RNAs exceeding a length of 200 nucleotides play an important role in ensuring cell functions and proper organism development by interacting with cellular compounds such as miRNA, mRNA, DNA and proteins. However, there is an additional level of lncRNA regulation, called lncRNA epigenetics, in gene expression control.
- lncRNA epigenetics
- RNA modyfiing enzymes
- detection modified nucleotides
1. General Remarks
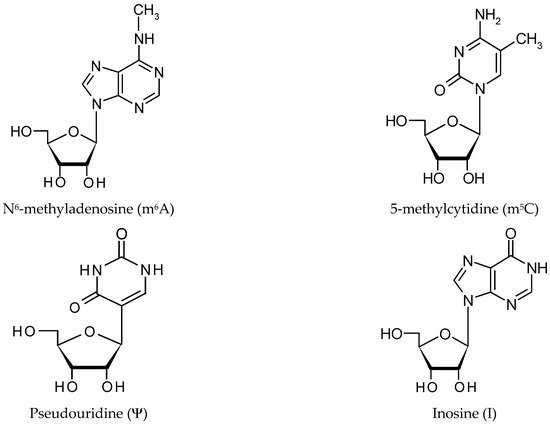
2. RNA Modification Mechanisms
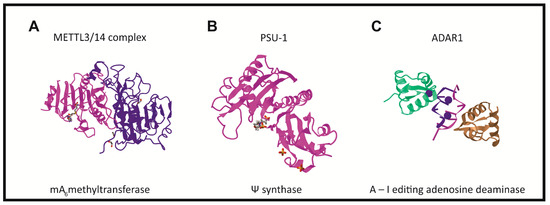
There are three groups of protein involved in modifying RNA metabolism [27,28][27][28]. The first group (writers,Figure 2) consists of enzymes introducing modified nucleotides into RNA during posttranscriptional RNA modifications; the second group of proteins interacts with modified nucleotides (readers); and the third group is involved in removing modification labels (erasers).
In humans, the formation of the m6A modification is connected with the methylase complex (writer). Crystallographic and biochemical studies have shown that METTL3 is S-adenosylmethionine methyltransferase with catalytic properties, while METTL14 serves as an RNA binding platform (Figure 2A) [48][41]. Recently, METTL16 was characterized as another “writer” protein, which interacts with several types of RNAs: mRNA, U6 and lncRNA [58,59,60,61][42][43][44][45]. Unlike METTL3/METTL14, which modifies A to m6A in coding RNAs, METTL16 can methylate both coding and noncoding RNAs [62][46].
Like DNA and histone modifications pathways, the m6A modification has two specific erasers, FTO and ALKBH5 [64,65][47][48]. FTO (fat mass and obesity-associated protein) removes the methylation trace by oxidizing m6A to N6-hydroxymethyladosine or N6-formyladenosine, which are chemically unstable and can hydrolyze to the final adenine product [66,67,68][49][50][51]. Another eraser is the homologous protein ALKBH5, which catalyzes the direct removal of the methyl group from adenine [69][52]. Whereas the FTO protein has been linked to obesity, the ALKBH5 protein is essential to spermatogenesis.
The human YTH domain containing protein family consists of five proteins, namely, YTHDF1–3 (eIF3) binds to the m6A located in the 5′UTR region of mRNA and is involved in cap-independent translation [82,83,84][56][57][58]. This mechanism is known as the “RNA epigenetic m6A switch”, which means m6A alters the local structure of mRNA or lncRNA, to facilitate the binding of HNRNPs for biological regulation [87][59].
Two writer m5C methyltransferases (MTases) have been shown to catalyze the m5C modification of eukaryotic RNA. One of them, RNA DNMT2, resembles DNA methyltransferases in its structure and characteristics [92[60][61],93], whereas the second group comprised of seven members, the MTPases (NSUN), contains the conserved NOL1/Nop2/Sun motif [94][62]. In 2017, Yang et al. presented evidence through in vitro and in vivo studies that m5C formation in mRNAs is mainly catalyzed by the NSUN2 type RNA methyltransferase [95][63]. Moreover, hm5C modifications of RNA are involved in stem cell pluripotency and impact translation efficiency [97,98][64][65].
[28]. Ψ has an unusual nucleoside, containing a C–C glycosidic bond, instead of the N-C bond found in the rest of the nucleosides [100][66]. As a result of U being replaced by Ψ, an additional hydrogen bond donor is present at the non-Watson–Crick edge. The distinct structure of Ψ increases both the rigidity of the phosphodiester backbone, as well as the thermodynamic stability of Ψ–A, compared with U–A [101,102][67][68].
Pseudouridine writers, called pseudouridine synthases (PUSs), recognize substrates and catalyze the isomerization of U to Ψ, without the need for cofactors (Figure 2B) [42,100,103][54][66][69]. However, PUS enzymes are unable to isomerize free nucleotides. In rRNA pseudouridylation, small nucleolar RNAs act as guides that recognize targets with sequence complementarity, thus directing pseudouridylation in a site-specific manner [104,105,106][70][71][72]. In the solved crystal structure, three of the proteins interact with the H/ACA guide RNA or substrate RNA, while GAR1 may regulate substrate loading and release [104,108][70][73].
In contrast to the m6A modification, specific Ψ eraser or reader proteins have not yet been identified. Furthermore, the lack of reader proteins that specifically bind to Ψ means it is difficult for proteins to identify the more subtle modification, which results in the C–N bond being replaced by a C–C bond, between uracil and ribose [109][74]. In humans, 10 proteins (PUS1–10) involved in RNA modification, with an annotated Ψ synthase domain, have been found (writers). It is also possible that there could be readers and erasers for the Ψ modification; however, their existence has not yet been proven.
ncRNAs and is catalyzed by the writer protein, adenosine deaminase acting on RNA (ADAR). ADAR1 and ADAR2 catalyze all the currently known A-to-I editing sites. Inosine essentially mimics the chemical properties of guanosine, therefore ADAR proteins introduce an A-to-G substitution in transcripts. These changes can lead to specific amino acid substitutions, altering protein composition.
3. Detection of Modified Nucleotides
The information presented above suggests that lncRNA epigenetics are important to cell differentiation and may be involved in controlling organism development. This has motivated many laboratories to screen ncRNAs for the presence of modified nucleotides, and to study the changes of the modification pattern during cell development [26,27][26][27].
Application NGS methods to epigenetic mapping is so far difficult because they typically do not detect modified nucleosides [116,117][75][76]. Developing single-base resolution sequencing, which could quantify the relatively low abundance of modified nucleotides in lncRNA, is a significant challenge. The identification of transcriptome-wide RNA modifications has been approached using different strategies.
The study of RNA modification started in 1957 when the first modified nucleoside, pseudouridine, was discovered in bulk yeast RNA, using paper chromatography [118][77]. The cut site is labeled with32P and the32P labeled RNA fragment is splint ligated to 116-nucleotide single stranded DNA oligonucleotide, using DNA ligase. The sample is then digested with RNase T1/A to completely digest all RNA, whereas the32Plabelled candidate site remains with the DNA nucleotide as DNA-32P(A/m6A)p and DNA-32P(A/m6A)Cp, which migrate as 117/116 mers on denaturing gel. This method was successfully used to determine the modified nucleotides like m6A, m5C, Ψ, and possibly other unknown modified nucleotides, in several coding and noncoding RNAs.
The high-throughput m6A mapping strategies were based on the immunoprecipitation of modified RNA molecules, using m6A-specific antibodies coupled to the subsequent NGS sequencing. Then, the RNA separated with a magnet is subjected to a second round of m6A immunoprecipitation. The resulting RNA pool, which is highly enriched with m6A-containing RNAs, is used for library construction and NGS sequencing [117,126][76][78]. miCLIP allows for a high-resolution detection of m6A in RNAs.
The detection of the m6A nucleotide in RNA, using an indirect approach, is difficult because few chemical reagents modify the methyl group. However, NOseq, a method for the detection of m6A in RNA after chemical deamination by nitrous acid, has recently been introduced [130][79]. Nitrous acid deaminates adenosines to inosine, while the m6A residue is resistant to such modifications. The application of NGS after modification to detect m6A sites in MALAT1 lncRNA
The most common indirect method used to determine m5C modification sites in RNA, i.e., the bisulfite conversion of cytidine to uridine has been applied successfully to determine the presence of this modified nucleotide in DNA [131][80]. The method is based on the fact that m5C modified cytosine is resistant to bisulfite cytosine deamination. There are several biochemical kits on the market, making it possible to prepare m5C libraries, which are ready for sequencing. Moreover, other modifications or double-stranded regions may be resistant to bisulfite treatment, especially under the milder reaction conditions required to maintain RNA integrity.
An alternative approach, which has been termed the “suicide enzyme trap”, has been employed to characterize the substrates of the following m5C-methyltransferases (m5C-MTases), NSUN2 and NSUN4 [137,138][81][82]. By mutating m5C-MTases to form irreversible covalent bonds with target residues, the resulting stable enzyme–RNA complexes are suitable for immunoprecipitation and mapping. This is also the case with the AZA-seq methodology, in which the “suicide inhibitor” nucleotide analog of 5-azacytidine is incorporated into cellular RNA and “traps” m5C-MTases for pulldown and sequencing [139][83].
Determining the Ψ sites in the RNA chain requires indirectly analyzing this modification using carbodiimide chemistry. The bulky CMCT group attached to N3 on Ψ hinders reverse transcription and results in cDNA being truncated. This facilitates the detection of Ψ at a single nucleotide resolution level [140][84]. Sites of pseudouridylation with single nucleotide resolution can be identified by subjecting the data obtained through NGS sequencing of Pseudo-seq libraries, to computational analysis.
Recently, Pan et al. developed a method that uses a CMC-Ψ-induced RT stop with an additional step of site-specific ligation, followed by PCR, to generate two unique PCR products, that correspond to the modified and unmodified uridine. The modification is visualized in the PCR products using gel electrophoresis [143][85].
As identifying true editing sites from transcriptome sequencing data is difficult, alternative methods aimed at marking inosine have been developed. RNase T1specifically cleaves RNA after guanosine or inosine but is inhibited by guanosine glyoxal/borate adducts. The cleavage of glyoxal-modified RNA creates RNA fragments that carry inosine at their termini, as an input for sequencing. ICE involves the treatment of RNA with acrylonitrile, which converts the inosine to N1-cyanoethylinosine in the process of cyanoethylation, and results in the formation of an inosine/acrylonitrile adduct that inhibits base pairing with cytidine and stalls reverse transcription.
Recently, a direct modification detection method, called nanopore sequencing, has been developed [155,156,157][86][87][88]. Tombo is a software used to detect modifications in DNA and RNA, such as the m5C modification in DNA and RNA and the m6A modification in DNA [160][89]. The EpiNano software is used to detect the m6A modification in RNA [117,155][76][86]. It needs to be highlighted that direct RNA modification analysis using nanopore sequencing is rapidly developing and becoming more reliable, so its routine application in the field of RNA epigenetics is expected.
4. The Impact of lncRNA Epigenetics on lncRNA Function
It is estimated that human cells have over 50,000 lncRNA molecules coded in genes. Many mature lncRNAs are modified after transcription (Section 2). The application of NGS methods in combination with bioinformatic analysis revealed the occurrence of several modifications in different types of lncRNA molecules. recruit factors, either to the site of lncRNA transcription or to adjacent loci and involve lncRNA XIST and lncRNA H19 [165,166][90][91].
Due to its size, XIST has many modifications like 78 sites m6A. 5 sites m5C and single site of Ψ [26]. In humans, multiple m6A sites in XIST repeat regions have been identified. mRNAs, is mediated by RBM15 and RBM15B, which bind the m6A-methylation complex and recruit it to specific sites in RNA [83][57]. Additionally, the knockdown of RBM15 and RBM15B, or the knockdown of METTL3 methyltransferase, impairs XIST-mediated gene silencing.
Many pathways contribute to the control of gene expression during development. Polycomb repressive complex (PRC2) and XIST are associated with gene repression in various developmental processes, such as X chromosome inactivation and genomic imprinting.
PRC2 binds with high affinity to the 5′-end region of XIST called the repeat In human XIST, five m5C marks were also detected. The presence of m5C methylation in the XIST transcript prevents the binding of the PRC2 in vitro. In Xist lncRNA, the presence of pseudouridylation and A–I editing sites has been confirmed, however, their roles are unknown at this stage so far [170,171][92][93].
The recent analysis of MeRIP seq data revealed thousands of m6A switches, which are involved in alternative RNA splicing and abundance [173][94]. They also show that the recognition of the m6A mark in MALAT1 by the YTHDC1 reader protein plays a critical role in maintaining the composition of nuclear speckles and their genomic binding sites. In addition,MALAT1lncRNA is subject to post-transcriptional m5C modification; five m5C sites been found to regulate chromatin-related roles in other lncRNAs, such as HOTAIR and XIST [135][95]. Although the exact role played by pseudouridine and inosine modifications remains to be explained, and the presence of each of the three inosines increases the stability of MALAT1 by 2–3 kcal/mol [28,170][28][92].
HOTAIR interacts with the nuclear m6A reader YTHDC1 at the methylated A783 and at additional sites [176][96]. Localization in chromatin strongly depends on the m6A modification at site A783 of HOTAIR, while the modification of other m6A sites mediates high HOTAIR levels. The previous results demonstrate that site-specific cytosine methylation occurs in lncRNA HOTAIR [135][95]. The methylation of C1683 is widespread in different cell types and it is not limited by the abundance of HOTAIR RNA levels.
This lincRNA is necessary for the differentiation of mouse embryonic stem cells (mESCs) and acts as a ceRNA by sequestering let-7 miRNAs [177][97]. It has been proposed that the presence of m6A in lincRNA1281 can act as a m6A-switch for specific RNA binding proteins, which will eventually regulate their interaction with let-7 miRNA. However, the identity of such proteins has not yet been discovered. A similar mechanism has already been proposed for the binding of the HuR (ELAVL1) protein and miRNAs, to the mRNAs encoding developmental regulators in mESCs.
H19, an imprinted lncRNA with a size of 2.3 kb, plays an important role in embryonic development [178][98]. The knockdown of METTL3 or METTL14 notably reversed the hypoxic preconditioning-induced (HPC-induced) enhancement of cell viability, anti-apoptosis ability, and lncRN AH19 expression [179][99]. The Ras-GTPase-activating protein-binding protein 1 (G3BP1) was confirmed to bind methylated lncRNAH19, based on the presence of NSUN2. lncRNA H19 has two editing sites, and their presence increases the lncRNA stabilization energy by 3 kcal/mol [171][93].
The human steroid receptor RNA activator (SRA) is a transcript of thesra1gene, whose size ranges from 0.7 to 0.9 kb. It is a dual-function RNA, which acts as both an lncRNA and an mRNA [181][100]. lncRNA SRA regulates several processes, such as cell cycle proliferation, as well as insulin, Notch, and TNFa signaling [182][101]. In a subsequent study, the same authors identified a specific uridine residue in SRA1 (U206), whose modification by PUS1 (or PUS3) might induce a functional switch, which regulates nuclear receptor signaling [26,181][26][100].
It serves an oncogenic role in a variety of malignant tumors, such as colorectal cancer [183][102]. PVT1 lncRNA is highly modified and contains m6A, m5C and Ψ Moreover, the RNA m6A modification mediated by the METTL3/METTL14 complex (Figure 2A) regulates epidermal stemness by controlling Pvt1 and MYC interactions through Pvt1 methylation, uncovering a key and novel molecular mechanism underlying skin tissue homeostasis, regeneration and wound repair. Some of the Ψ sites were located within functional lncRNA motifs, indicating the potential regulatory impact of Ψ on lncRNAs.
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Xue, C. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Kapranov, P.; Willingham, A.T.; Gingeras, T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007, 8, 413–423.
- Layton, E.; Fairhurst, A.M.; Griffiths-Jones, S.; Grencis, R.K.; Roberts, I.S. Regulatory RNAs: A universal language for inter-domain communication. Int. J. Mol. Sci. 2020, 21, 8919.
- Uchida, S.V.; Adams, J.C. Physiological roles of non-coding RNAs. Am. J. Physiol. Cell 2019, 317, C1–C2.
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2017, 17, 47–62.
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The landscape of long non-coding RNA classification. Trends Genet. 2015, 31, 239–251.
- Schulz, D.; Schwalb, B.; Kiesel, A.; Baejen, C.; Torkler, P.; Gagneur, J.; Soeding, J.; Cramer, P. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 2013, 155, 1075–1087.
- Zhu, S.; Wang, J.; He, Y.; Meng, N.; Yan, G.R. Peptides/Proteins encoded by non-coding RNA: A novel resource bank for drug targets and biomarkers. Front. Pharmacol. 2018, 9, 1295.
- Chugunova, A.; Navalayeu, T.; Dontsova, O.; Sergiev, P. Mining for small translated ORFs. J. Proteome Res. 2018, 17, 1–11.
- Matsumoto, A.; Nakayama, K.I. Hidden peptides encoded by putative noncoding RNAs. Cell Struct. Funct. 2018, 43, 75–83.
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human long noncoding RNA interactome: Detection, characterization and function. Int. J. Mol. Sci. 2020, 21, 1027.
- Rinn, J.L.; Chang, H.Y. Long noncoding RNAs: Molecular modalities to organismal functions. Annu. Rev. Biochem. 2020, 89, 283–308.
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573.
- Roundtree, I.A.; Molly, E.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200.
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev Genet. 2008, 9, 465–476.
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322.
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 2018, 9, 640.
- Carter, C.; Zhao, K. The epigenetic basis of cellular heterogenity. Nat. Rev. Genet. 2020, 169, 1187–1200.
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307.
- Itaya, T.; Kanai, T.; Sawada, T. Structure of wyosine, the condensed tricyclic nucleoside of torula yeast phenylalanine transfer ribonucleic acid. Chem. Pharm. Bull. 2002, 50, 547–548.
- Grosjean, H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution; Landes Bioscience: Austin, TX, USA; CRC Press: Boca Raton, FL, USA, 2009.
- Ajitkumar, P.; Cherayil, J.D. Thionucleosides in transfer ribonucleic acid: Diversity, structure, biosynthesis, and function. Microbiol. Rev. 1988, 52, 103–113.
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 2017, 14, 1197–1208.
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539.
- Jacob, R.; Zander, S.; Gutschner, T. The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 2017, 18, 2387.
- Dinescu, S.; Ignat, S.; Lazar, A.D.; Constantin, C.; Neagu, M.; Costache, M. Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes 2019, 10, 52.
- De Zoysa, M.D.; Yu, Y.T. Posttranscriptional RNA pseudouridylation. Enzymes 2017, 41, 151–167.
- Torres, A.G.; Pineyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286.
- Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018, 28, 1–10.
- Lyons, S.M.; Fay, M.M.; Ivanov, P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS J. 2018, 592, 2828–2844.
- Avcilar-Kucukgoze, I.; Kashina, A. Hijacking tRNAs from translation: Regulatory functions of tRNAs in mammalian cell physiology. Front. Mol. Biosci. 2020, 7, 610–617.
- Sylvers, L.A.; Rogers, K.C.; Shimizu, M.; Othsuka, E.; Soll, D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 1993, 32, 3836–3841.
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408.
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840.
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2—Mediated m5C methylation and METTL3/METTL14—mediated m6A methylation cooperatively enhance p21 translation. J. Cell Biochem. 2017, 118, 2587–2598.
- Hoernes, T.P.; Faserl, K.; Juen, M.A.; Kremser, J.; Gasser, C.; Fuchs, E.; Shi, X.; Siewert, A.; Lindner, H.; Kreutz, C.; et al. Translation of non-standard codon nucleotides reveals minimal requirements for codon-anticodon interactions. Nat. Commun. 2018, 9, 4865.
- Sloan, K.E.; Warda, A.S.; Sharma, K.E.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152.
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351.
- Popis, M.C.; Blanco, S.; Frye, M. Posttranscriptional methylation of transfer and ribosomal RNA in stress response pathways, cell differentiation, and cancer. Curr. Opin. Oncol. 2016, 28, 65–71.
- Liu, P.; Li, F.; Lin, J.; Fukumoto, T.; Nacarelli, T.; Hao, X.; Kossenkov, A.V.; Simon, M.C.; Zhang, R. m6A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat. Cell Biol. 2021, 23, 355–365.
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Markus, T.; Bohnsack, M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014.
- Ruszkowska, A.; Ruszkowski, M.; Dauter, Z.; Brown, J.A. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018, 8, 53.
- Mendel, M.; Chen, K.M.; Homolka, D.; Gos, P.; Pandey, R.R.; McCarthy, A.A.; Pillai, R.S. Methylation of structured RNA by the m6A writer METTL16 Is essential for mouse embryonic development. Mol. Cell 2018, 71, 986–1000.e11.
- Ruszkowska, A. METTL16, methyltransferase-like protein 16: Current insights into structure and function. Int. J. Mol. Sci. 2021, 22, 2176.
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 2017, 168, 824–835.
- Nance, D.J.; Satterwhite, E.R.; Bhaskar, B.; Misra, S.; Carraway, K.R.; Mansfield, K.D. Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS ONE 2020, 15, e0227647.
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624.
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014, 15, 203–306.
- Gerken, T.; Girard, C.A.; Tung, Y.C.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.; McDonough, M.A. The obesity-associated FTO gene encodes a 2-oxoglutaratedependent nucleic acid demethylase. Science 2007, 318, 1469–1472.
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887.
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013, 49, 18–29.
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. METTL3/METTL14 m6A methyltransferase. Nature 2016, 534, 575–578.
- Czudnochowski, N.; Wang, A.L.; Finer-Moore, J.; Stroud, R.M.J. In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. Mol. Biol. 2013, 425, 3875–3887.
- Placido, D.; Brown, B.A.; Lowenhaupt, K.; Rich, A.; Athanasiadis, A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure 2007, 15, 395–404.
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A promotes cap-independent translation. Cell 2015, 164, 999–1010.
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m 6 A in the transcriptome: M 6 A-binding proteins. Trends Cell Biol. 2018, 28, 113–127.
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831.
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564.
- Jeltsch, A.; Nellen, W.; Lyko, F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biol. Sci. 2006, 31, 306–308.
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Mark Helm, M. Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA Biol. 2017, 14, 1108–1123.
- Chi, L.; Delgado-Olguín, P. Expression of NOL1/NOP2/sun domain (Nsun) RNA methyl- transferase family genes in early mouse embryogenesis. Gene Expres. Patterns 2013, 13, 319–327.
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625.
- Yang, J.; Bashkenova, N.; Zang, R.; Huang, X.; Wang, J. The roles of TET family proteins in development and stem cells. Development 2020, 147, dev183129.
- Shen, H.; Ontiveros, R.J.; Owen, M.C.; Liu, M.Y.; Ghanty, U.; Kohli, R.M.; Liu, K.F. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem. 2021, 296, 100087.
- Gray, M.C.; Charette, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351.
- Kierzek, E.; Malgowska, M.; Lisowiec, J.; Turner, D.H.; Gdaniec, Z.; Kierzek, R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014, 42, 3492–3501.
- Deb, I.; Popenda, Ł.; Sarzyńska, J.; Małgowska, M.; Lahiri, A.; Gdaniec, Z.; Kierzek, R. Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair. Sci. Rep. 2019, 9, 16278.
- Borchardt, E.K.; Martinez, N.M.; Gilbert, W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020, 54, 309–336.
- Zhao, Y.; Dunker, V.; Yu, Y.-T.; Karijolich, J. The Role of Noncoding RNA Pseudouridylation in Nuclear Gene Expression Events. Front. Bioeng. Biotechnol. 2018, 6, 8.
- Ni, J.; Tien, A.L.; Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997, 89, 565–573.
- Caton, E.A.; Kelly, E.K.; Kamalampeta, R.; Kothe, U. Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 2018, 46, 905–916.
- Li, X.; Ye, K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 2006, 443, 302–307.
- Li, X.; Ma, S.; Yi, C. Pseudouridine: The fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 2016, 33, 108–116.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Liu, H.; Begik, O.; Lucas, M.C.; Ramirez, J.M.; Mason, C.E.; Wiener, D.; Schwartz, S.; Mattick, J.S.; Smith, M.A.; Novoa, E.M. Accurate detection of m6 RNA modifications in native RNA sequence. Nat. Commun. 2019, 10, 4079.
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957, 227, 907–915.
- Zhang, C.; Fu, J.; Zhou, Y. A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 2019, 10, 922.
- Werner, S.; Galliot, A.; Pichot, F.; Kemmer, T.; Marchand, V.; Sednev, M.V.; Lence, T.; Roignant, J.Y.; König, J.; Höbartner, C.; et al. NOseq: Amplicon sequencing evaluation method for RNA m6A sites after chemical deamination. Nucleic Acids Res. 2021, 49, e23.
- Clark, S.J.; Frommer, M. Bisulfite genomic sequencing of methylated cytosines. In Laboratory Methods for the Detection of Mutations and Polymorphisms in DNA; Taylor, G., Ed.; CRC Press: New York, NY, USA, 1997; pp. 151–162.
- Metodiev, M.D.; Spåhr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.G.; Ruzzenente, B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110.
- Khoddami, V.; Cairns, B. Transcriptome—Wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat. Protoc. 2014, 9, 337–361.
- Uddin, M.B.; Zhishan Wang, Z.; Yang, C. Dysregulations of functional RNA modifications in cancer, cancer stemness and cancer therapeutics. Theranostic 2020, 10, 3164–3189.
- Ofengand, J.; Bakin, A.; Wrzesinski, J.; Nurse, K.; Lane, B.G. The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol. 1995, 73, 915–924.
- Zhang, W.; Eckwahl, M.J.; Zhou, K.I.; Pan, T. Sensitive and quantitative probing of pseudouridine modification in mRNA and long noncoding RNA. RNA 2019, 25, 1218–1225.
- Xu, L.; Seki, M. Recent advances in the detection of base modifications using the Nanopore sequencer. J. Hum. Genet. 2020, 65, 25–33.
- Motorin, Y.; Marchand, V. Analysis of RNA modifications by second- and third-generation deep sequencing: 2020 Update. Genes 2021, 12, 278.
- Furlan, M.; Tanaka, I.; Leonardi, T.; de Pretis, S.; Pelizzola, M. Direct RNA sequencing for the study of synthesis, processing, and degradation of modified transcripts. Front. Genet. 2020, 11, 394.
- Jia, L.; Chen, J.; Liu, H.; Fan, W.; Wang, D.; Li, J.; Liu, D. Potential m6A and m5C methylations within the genome of a chinese african swine fever virus strain. J. Virol. Sin. 2020, 36, 321–324.
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407.
- Zhao, Y.; Teng, H.; Yao, F.; Yap, S.; Sun, Y.; Ma, L. Challenges and Strategies in Ascribing Functions to Long Noncoding RNAs. Cancers 2020, 12, 1458.
- Trotman, J.B.; Braceros, K.C.A.; Cherney, R.E.; Murvin, M.M.; Calabrese, J.M. The control of polycomb repressive complexes by long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2021, e1657.
- LNCipedia/NONCODE. Available online: (accessed on 1 June 2021).
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063.
- Amort, T.; Soulière, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Wörle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008.
- Porman, A.M.; Roberts, J.R.; Chrupcala, M.; Kennedy, M.; Williams, M.M.; Richer, J.K.; Johnson, A.M. A single N6-methyladenosine site in lncRNA HOTAIR regulates its function in breast cancer cells. BioRaiv 2020.
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920.
- Gabory, A.; Dandolo, L.; Yoshimizu, T.; Ripoche, M.A. The H19 gene: Regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006, 113, 188–193.
- Su, Y.; Xu, R.; Zhang, R.; Qu, Y.; Zuo, W.; Ji, Z.; Geng, H.; Pan, M.; Ma, G. N6-methyladenosine methyltransferase plays a role in hypoxic preconditioning partially through the interaction with lncRNA H19. Acta Biochim. Biophys. Sin. 2020, 52, 1306–1315.
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27.
- Sheng, L.; Ye, L.; Zhang, D.; Cawthorn, W.P.; Xu, B. New Insights Into the Long Non-coding RNA SRA: Physiological Functions and Mechanisms of Action. Front. Med. 2018, 5, 244.
- Jin, K.; Wang, S.; Zhang, Y.; Xia, M.; Mo, Y.; Li, X.; Li, G.; Zeng, Z.; Xiong, W.; He, Y. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol. Life Sci. 2019, 76, 4275–4289.
