Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Xiangru Wang and Version 1 by Xiangru Wang.
Brain microvascular endothelial cells (BMECs) constitute the structural and functional basis for the blood–brain barrier (BBB) and play essential roles in bacterial meningitis. Electrical cell-substrate impedance sensing (ECIS) measurement and Western blot assay demonstrated lncRSPH9-4 overexpression in hBMECs mediated the BBB integrity disruption.
- brain microvascular endothelial cells
- lncRSPH9-4
- miR-17-5p
- MMP3
- tight junctions
1. Introduction
Bacterial meningitis is the most important life-threatening infection of the central nervous system (CNS) with high morbidity and mortality and Escherichia coli is the most common gram-negative pathogenic bacterium causing this outcome [1]. Most bacterial meningitis develops from bacterial penetration of the BBB [2]. Vascular endothelium constitutes the structural and functional basis of the BBB and plays an important role in maintaining the integrity of the BBB, as well as CNS homeostasis [3]. BMECs are characterized by the presence of tight junction proteins (TJs), including Claudins, Occludin and zonula-occludens [4,5][4][5]. BMECs dysfunction is often caused by the decrease or re-distribution of these TJs, which lead to disruption of the BBB [6]. Thus, protecting and maintaining the BBB function is of positive significance in alleviating brain damage after the CNS-invading pathogens infection.
LncRNAs are considered regulators of diverse biological processes, including imprinting control, cell differentiation and development [7[7][8][9],8,9], as well as many pathological processes, such as cancer, chronic inflammation and infectious diseases [10,11,12][10][11][12]. Aberrant expression and mutations of lncRNAs uncovered in various brain dysfunctions have led researchers to investigate the potential roles of lncRNAs in brain physiology and pathology [13]. For example, lncRNAs MALAT1 GAS5 and NEAT1 are widely recognized to be implicated in cancer, vascular diseases and neurological disorders [14,15,16,17][14][15][16][17]. MALAT1 was reported to promote the BMEC autophagy; NEAT1 was able to activate the NF-κB signaling pathway in nerve cells [17,18][17][18]. Regarding lncRNAs, the competing endogenous RNA (ceRNA) working mechanism has been largely evidenced, in which lncRNAs act as the ceRNA to interact competitively with miRNAs and regulate the expression of the target proteins [19]. Our previous lncRNA transcriptomics sequencing identified a batch of significantly different lncRNAs in hBMECs upon meningitic E. coli infection [20] and we further noticed one novel lncRNA, lncRSPH9-4, that was significantly up-regulated along with the infection. We focused on this lncRNA because we found that overexpressing lncRSPH9-4 in hBMECs would cause a decrease of the impedance values. However, the specific working mechanism of this lncRNA in the pathogenic process is unclear. A variety of miRNAs have been reported to regulate the integrity of the BBB, such as miR-18a, miR-338-3p and miR-182 [13,21,22][13][21][22].
2. Discussion
Due to their extensive involvement in biological processes, lncRNAs have attracted immense research interest in recent years. LncRNAs have been increasingly involved in the regulation of multiple central nervous system disorders, such as ischemic stroke, multiple sclerosis and Huntington’s disease [16,27,28][16][23][24]. However, in CNS infectious diseases, the regulatory function of lncRNAs was largely unclear. In this study, we identified and characterized an up-regulated lncRNA, lncRSPH9-4, in hBMECs challenged with meningitic E. coli. We demonstrated that lncRSPH9-4 helped the infection-caused disruption of the BBB integrity via sponging miR-17-5p, thus promoted the expression of MMP3 and eventually increased the degradation of TJs. Lots of lncRNAs were involved in the development of neurodegenerative diseases. A well-studied lncRNA, MALAT1, was reported to contribute to protecting the BBB after stroke [29][25]. Other lncRNAs, such as BC200 and Sox2OT, were found to be associated with Alzheimer’s disease or Parkinson’s disease [30,31][26][27]. Accumulating pieces of evidence have also supported the essential roles of lncRNAs in virus infection. For example, a lncRNA called lncRNA-ACOD1 was found to facilitate Vesicular vtomatitis virus replication in macrophages through enhancing GOT2 enzymatic activity [12]. LncRNAs exerted their functions from many aspects. In recent years, the concept of ceRNA as a new regulatory mechanism has been raised and widely accepted in diverse physiological and pathological processes, which means that lncRNAs could act as a ceRNA to competitively sponge miRNA, thus resulting in a decreased mRNA degradation [32][28]. Numerous studies have increasingly reported that lncRNAs function as sponges to interact with miRNAs at the post-transcriptional level [33,34][29][30]. For example, a lncRNA named linc-EPHA6-1 can sponge miR-4485-5p to regulate NKP46 expression and enhance NK cells cytotoxicity against Zika virus-infected cells [35][31]. However, there are few studies about the lncRNAs function in bacterial infectious diseases. We previously found one lncRNA, lncRSPH9-4, that was significantly increased in meningitic E. coli-infected hBMECs. Here, we further analyzed the transcriptomic profiles in hBMECs in response to the overexpression of lncRSPH9-4 via RNA-sequencing. A total of 639 mRNAs and 299 miRNAs with significant alteration upon lncRSPH9-4 overexpression have been identified and the ceRNA regulatory network was built based on the sequencing data. One miRNA, miR-17-5p, showed the possibility of being sponged by lncRSPH9-4 and influenced the expression of MMP3, an important protein associated with intercellular integrity and permeability. It is worth wondering about the relationship between miR-17-5p and the BBB permeability. The BBB is ultrastructurally assembled by a monolayer of BMECs, which are tightly attached via TJs and adherens junctions (AJs) [6,36][6][32]. Many miRNAs have been shown to influence the BBB permeability. Some pathogens, such as Coxsackievirus A16, can increase the expression of miRNA-1303 and trigger the changes in the BBB permeability [37][33]. Other miRNAs, such as miRNA-29b and miRNA-15, were reported to control the BBB integrity by targeting MMP9 [38,39][34][35]. Exosomes-derived miRNA-132 regulated the expression of vascular endothelial cadherin (VE-cadherin) by directly targeting eukaryotic elongation factor 2 kinase (eef2k) and increased the permeability of the BBB [40][36]. It has been reported that miR-17-5p plays a vital role in endothelial cells by binding to VEGF-A 3′UTR and inhibiting VEGF-A expression in HUVECs [41][37]. Here, in our study, we used an immortalized hBMECs cell line, established by Kwang Sik Kim in Johns Hopkins University School of Medicine, as our in vitro BBB model. This immortalized hBMECs line was positive for Factor VIII-Rag, could intake DiI-AcLDL and showed a positive reaction for GGTP, which indicated that they exhibited brain endothelial characteristics [42][38]. It also expressed VCAM-1, ICAM-1 and TJs and maintained their barrier properties [43][39]. The hBMECs monolayer model provided by Kim has been largely applied in BBB functional research. For example, molecules such as CC chemokine ligand 2 (CCL2) and snail family transcriptional repressor 1 (Snail1) have been reported to contribute to blood–brain barrier disruption by the application of the hBMEC monolayer [44,45][40][41]. In our study, we found that miR-17-5p expression was significantly downregulated in hBMECs when overexpressing lncRSPH9-4. Consistently, miR-17-5p was also decreased in hBMECs in response to meningitic E. coli infection, as observed in our previous work [46][42]. Here, we showed that miR-17-5p was one of the competitive-sponging targets of lncRSPH9-4 and miR-17-5p could bind to 3′UTR of MMP3 to suppress the expression of MMP3. MMP3 is a member of the MMP family, which is widely involved in the breakdown of extracellular matrix proteins during tissue remodeling, such as embryonic development and reproduction, as well as in disease processes, such as arthritis and tumor metastasis [47][43]. MMP3 can degrade collagen, elastin, fibronectin, laminin, as well as tight junctions, so as to increase vascular permeability and disrupt the BBB [25,41,48,49][44][37][45][46]. In hBMECs, our results suggested that lncRSPH9-4 was able to regulate MMP3 expression by acting as a competitive sponge of miR-17-5p. Since previous studies have already evidenced that MMP3 can directly degrade the TJs of vascular endothelial cells [25[44][37],41], we therefore next investigated the potential effects on MMP3 regulated by lncRSPH9-4 and miR-17-5p and results showed that lncRSPH9-4 could negatively regulate the TJs, while miR-17-5p positively regulated the TJs and, importantly, this regulatory axis was probably mediated by the change of MMP3 expression during this process. Taken together, our findings provided substantial evidence, for the first time clarifying the regulatory function of lncRNA during meningitis-causing bacterial infection. We demonstrated that meningitic E. coli-induced lncRSPH9-4 could function as a ceRNA to compete with miR-17-5p, which led to the increased expression of MMP3 and therefore the degradation of the TJs barrier (Figure 1). However, the mechanisms of BBB disruption in the development of bacterial meningitis are quite complex and the contribution of lncRSPH9-4 is just the tip of the iceberg. At the same time, we investigated the expression specificity of lncRSPH9-4 in the other endothelial cells (peripheric endothelial cells, such as HUVECs), as well as other barrier-forming (not endothelium) cells, such as astrocytes, in our previous work. As demonstrated below, lncRSPH9-4 was also significantly induced by the challenge of meningitic E. coli, indicating that lncRSPH9-4 probably exhibits a similar working mechanism in the astrocytes cell line U251. We also noticed the expression/transcription of lncRSPH9-4 in the peripheric vascular endothelial cells HUVECs; however, it was not significantly induced by the infection, suggesting that lncRSPH9-4 might not work in this type of endothelial cells [20]. At last, further efforts are required to explore the generation of lncRSPH9-4 and whether there are some other targets as well as regulative mechanisms of lncRSPH9-4 in meningitic bacterial penetration of the BBB.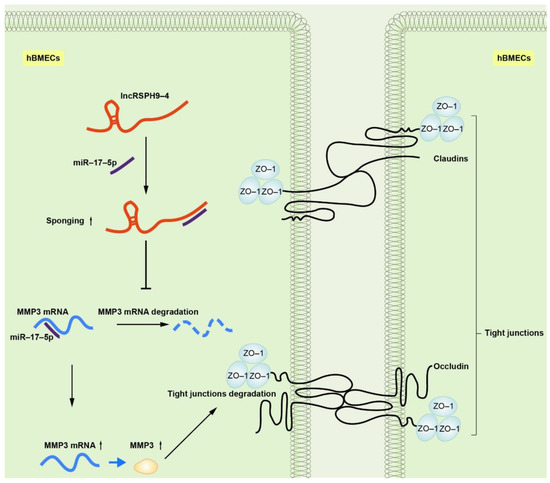
Figure 1. Schematic of the working mechanism of lncRSPH9-4 in the regulation of the endothelial barrier disruption. Meningitic E. coli infection of hBMECs induced the up-regulation of lncRSPH9-4, which could sponge miR-17-5p and thus increase the expression of MMP3 in hBMECs and finally lead to the tight junction degradation.
3. Conclusions
We demonstrated that meningitic E. coli infection of hBMECs induced the significant upregulation of lncRSPH9-4, which facilitated the barrier disruption of the endothelial cells, probably through lncRSPH9-4/miR-17-5p/MMP3 axis. This finding provides a new idea for understanding the function of lncRNAs in bacterial infection, especially in bacterial penetration of the CNS. Blocking these lncRNAs, such as lncRSPH9-4, may also represent a new strategy to better prevent and improve the CNS dysfunctions.
References
- Kim, K.S. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 2008, 6, 625–634.
- Kim, K.S. Pathogenesis of bacterial meningitis: From bacteraemia to neuronal injury. Nat. Rev. Neurosci. 2003, 4, 376–385.
- Sandoval, K.E.; Witt, K.A. Blood-brain barrier tight junction permeability and ischemic stroke. Neurobiol. Dis. 2008, 32, 200–219.
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell 2009, 16, 209–221.
- Edwards, V.L.; Wang, L.C.; Dawson, V.; Stein, D.C.; Song, W. Neisseria gonorrhoeae breaches the apical junction of polarized epithelial cells for transmigration by activating EGFR. Cell. Microbiol. 2013, 15, 1042–1057.
- Hawkins, B.T.; Davis, T.P. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 2005, 57, 173–185.
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307.
- Lee, J.T.; Bartolomei, M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013, 152, 1308–1323.
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21.
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 2010, 19, R152–R161.
- Huang, W.; Thomas, B.; Flynn, R.A.; Gavzy, S.J.; Wu, L.; Kim, S.V.; Hall, J.A.; Miraldi, E.R.; Ng, C.P.; Rigo, F.; et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 2015, 528, 517–522.
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055.
- Zhang, L.; Luo, X.; Chen, F.; Yuan, W.; Xiao, X.; Zhang, X.; Dong, Y.; Zhang, Y.; Liu, Y. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J. Cell. Biochem. 2018, 119, 5460–5472.
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041.
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015, 121, 101–108.
- Sun, D.; Yu, Z.; Fang, X.; Liu, M.; Pu, Y.; Shao, Q.; Wang, D.; Zhao, X.; Huang, A.; Xiang, Z.; et al. LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017, 18, 1801–1816.
- Liu, W.Q.; Wang, Y.J.; Zheng, Y.; Chen, X. Effects of long non-coding RNA NEAT1 on sepsis-induced brain injury in mice via NF-kappaB. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3933–3939.
- Li, Z.; Li, J.; Tang, N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017, 354, 1–10.
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352.
- Yang, R.; Huang, F.; Fu, J.; Dou, B.; Xu, B.; Miao, L.; Liu, W.; Yang, X.; Tan, C.; Chen, H.; et al. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016, 6, 38903.
- Yang, X.; Zi, X.H. LncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1alpha axis. Brain Res. 2019, 1714, 174–181.
- Zhang, T.; Tian, C.; Wu, J.; Zhang, Y.; Wang, J.; Kong, Q.; Mu, L.; Sun, B.; Ai, T.; Wang, Y.; et al. MicroRNA-182 exacerbates blood-brain barrier (BBB) disruption by downregulating the mTOR/FOXO1 pathway in cerebral ischemia. FASEB J. 2020.
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254.
- Bao, M.H.; Szeto, V.; Yang, B.B.; Zhu, S.Z.; Sun, H.S.; Feng, Z.P. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018, 9, 281.
- Ruan, W.; Li, J.; Xu, Y.; Wang, Y.; Zhao, F.; Yang, X.; Jiang, H.; Zhang, L.; Saavedra, J.M.; Shi, L.; et al. MALAT1 Up-Regulator Polydatin Protects Brain Microvascular Integrity and Ameliorates Stroke Through C/EBPbeta/MALAT1/CREB/PGC-1alpha/PPARgamma Pathway. Cell. Mol. Neurobiol. 2019, 39, 265–286.
- Mus, E.; Hof, P.R.; Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 10679–10684.
- Arisi, I.; D’Onofrio, M.; Brandi, R.; Felsani, A.; Capsoni, S.; Drovandi, G.; Felici, G.; Weitschek, E.; Bertolazzi, P.; Cattaneo, A. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: Mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 2011, 24, 721–738.
- Yue, X.H.; Guo, L.; Wang, Z.Y.; Jia, T.H. Inhibition of miR-17-5p promotes mesenchymal stem cells to repair spinal cord injury. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3899–3907.
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358.
- Han, X.; Yang, F.; Cao, H.; Liang, Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015, 29, 3054–3064.
- Li, S.; Zhu, A.; Ren, K.; Li, S.; Chen, L. IFNbeta-induced exosomal linc-EPHA6-1 promotes cytotoxicity of NK cells by acting as a ceRNA for hsa-miR-4485-5p to up-regulate NKp46 expression. Life Sci. 2020, 257, 118064.
- Hermann, D.M.; ElAli, A. The abluminal endothelial membrane in neurovascular remodeling in health and disease. Sci. Signal 2012, 5, re4.
- Song, J.; Hu, Y.; Li, H.; Huang, X.; Zheng, H.; Hu, Y.; Wang, J.; Jiang, X.; Li, J.; Yang, Z.; et al. miR-1303 regulates BBB permeability and promotes CNS lesions following CA16 infections by directly targeting MMP9. Emerg. Microbes Infect. 2018, 7, 155.
- Deng, X.; Zhong, Y.; Gu, L.; Shen, W.; Guo, J. MiR-21 involve in ERK-mediated upregulation of MMP9 in the rat hippocampus following cerebral ischemia. Brain Res. Bull. 2013, 94, 56–62.
- Kalani, A.; Kamat, P.K.; Familtseva, A.; Chaturvedi, P.; Muradashvili, N.; Narayanan, N.; Tyagi, S.C.; Tyagi, N. Role of microRNA29b in blood-brain barrier dysfunction during hyperhomocysteinemia: An epigenetic mechanism. J. Cereb. Blood Flow Metab. 2014, 34, 1212–1222.
- Xu, B.; Zhang, Y.; Du, X.F.; Li, J.; Zi, H.X.; Bu, J.W.; Yan, Y.; Han, H.; Du, J.L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell. Res. 2017, 27, 882–897.
- Yang, Y.; Kimura-Ohba, S.; Thompson, J.F.; Salayandia, V.M.; Cosse, M.; Raz, L.; Jalal, F.Y.; Rosenberg, G.A. Vascular tight junction disruption and angiogenesis in spontaneously hypertensive rat with neuroinflammatory white matter injury. Neurobiol. Dis. 2018, 114, 95–110.
- Stins, M.F.; Badger, J.; Sik Kim, K. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb. Pathog. 2001, 30, 19–28.
- Stins, M.F.; Gilles, F.; Kim, K.S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J. Neuroimmunol. 1997, 76, 81–90.
- Kim, B.J.; Hancock, B.M.; Bermudez, A.; Del Cid, N.; Reyes, E.; van Sorge, N.M.; Lauth, X.; Smurthwaite, C.A.; Hilton, B.J.; Stotland, A.; et al. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J. Clin. Investig. 2015, 125, 2473–2483.
- Liu, T.; Miao, Z.; Jiang, J.; Yuan, S.; Fang, W.; Li, B.; Chen, Y. Visfatin Mediates SCLC Cells Migration across Brain Endothelial Cells through Upregulation of CCL2. Int. J. Mol. Sci. 2015, 16, 11439–11451.
- Yang, R.; Chen, J.; Xu, B.; Yang, B.; Fu, J.; Xiao, S.; Tan, C.; Chen, H.; Wang, X. circ_2858 Helps Blood-Brain Barrier Disruption by Increasing VEGFA via Sponging miR-93-5p during Escherichia coli Meningitis. Mol. Ther. Nucleic Acids 2020, 22, 708–721.
- Lerner, A.; Neidhofer, S.; Reuter, S.; Matthias, T. MMP3 is a reliable marker for disease activity, radiological monitoring, disease outcome predictability, and therapeutic response in rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2018, 32, 550–562.
- Gurney, K.J.; Estrada, E.Y.; Rosenberg, G.A. Blood-brain barrier disruption by stromelysin-1 facilitates neutrophil infiltration in neuroinflammation. Neurobiol. Dis. 2006, 23, 87–96.
- Zhang, Q.; Zheng, M.; Betancourt, C.E.; Liu, L.; Sitikov, A.; Sladojevic, N.; Zhao, Q.; Zhang, J.H.; Liao, J.K.; Wu, R. Increase in Blood-Brain Barrier (BBB) Permeability Is Regulated by MMP3 via the ERK Signaling Pathway. Oxid. Med. Cell Longev. 2021, 2021, 6655122.
- Rosenberg, G.A.; Yang, Y. Vasogenic edema due to tight junction disruption by matrix metalloproteinases in cerebral ischemia. Neurosurg. Focus 2007, 22, E4.
More
