Multiple biologically active components of human milk support infant growth, health and development. Milk provides a wide spectrum of mammary epithelial cell-derived extracellular vesicles (MEVs) for the infant. Although the whole spectrum of MEVs appears to be of functional importance for the growing infant, the majority of recent studies report on the MEV subfraction of milk exosomes (MEX) and their miRNA cargo, which are in the focus of this entry.
- adipogenesis
- DNA methyltransferase 1
- immune tolerance
- intestinal maturation
- milk exosome
- milk miRNAs
- necrotizing enterocolitis
- nuclear factor-κB
- receptor-interacting protein 140
- systemic milk effects
1. Introduction
Breastfeeding is considered to represent the ideal source of infant nutrition. During the postnatal period, the infant´s epithelial barrier of the gastrointestinal (GI) tract matures, while adaptive immunity is still developing [1]. Accumulating evidence indicates that human milk (HM) is critically involved in the regulation of intestinal maturation and immune cell education [2,3][2][3]. Multiple biologically active components of HM and various interacting signaling pathways drive developmental processes which remain largely obscure [4].
Recently, attention has been paid to the wide spectrum of lipid bilayer-enclosed milk extracellular vesicles (MEVs), especially the subfraction of milk exosomes (MEX) that contain proteins, lipids, Our perception that milk is not “just food” for the growing infant but represents a complex metabolic and endocrine signaling system for postnatal growth and programming via transfer of mTORC1-activating amino acids and gene-regulatory miRNAs [5,6,7][5][6][7] has been substantiated in recent years. MEX, a most important subfamily of MEVs, are biomolecular nanostructures released from mammary gland epithelial cells (MGECs), carrying specific biomolecular information. MEX are found in the milk of all mammals including HM and have received increasing scientific attention in recent years
miRNA-148a is also the most abundant miRNA of triacylglycerol-rich milk fat globules (MFGs) of HM [23][8]. Remarkably, the abundantly expressed miRNAs of human MEX exhibit striking nucleotide sequence homologies with the corresponding milk miRNAs of other mammals [20,25][9][10]. It has recently been demonstrated that the top 10 highly expressed MEX-derived miRNAs are evolutionarily conserved across the milk of various mammalian species, including humans [26][11]. Notably, the immune-related miRNAs enriched in MEX are resistant to harsh environmental conditions [32][12].
It is the intention of this review to provide up-to-date information on the impact of MEX and MEX-derived miRNAs on intestinal maturation and their systemic effects in human and animal tissues, which are important to understand the eminent role of MEX in infant health and development.
2. Exosomal miRNAs and Intestinal Maturation
Cells take up exosomes by a variety of endocytic pathways, including clathrin-dependent endocytosis, and clathrin-independent pathways such as caveolin-mediated uptake, macropinocytosis, phagocytosis and lipid raft-mediated internalization [43,44,45][13][14][15]. Bovine MEX uptake in human and rat intestinal epithelial cells (IECs) is mediated by endocytosis and depends on cell and exosome surface glycoproteins [46][16]. [20][9] demonstrated that incubation of human MEX with normal colon cells (CRL1831) significantly increased cellular levels of miRNA-148a and decreased the expression of DNMT1. Furthermore, the addition of human MEX to normal fetal colon epithelial cells increased cell proliferation in an miRNA-dependent manner [27][17].
Of note, knockdown of miRNA-148a inhibits IEC proliferation associated with an increase in the expression of DNMT1 [27][17]. It has been demonstrated in cultured human colonic LS174T cells that exposure to bovine MEX enhances the expression of glucose-regulated protein 94 (GRP94) Furthermore, GRP94 plays a crucial role in gut homeostasis via chaperoning crucial components of the canonical WNT pathway [50][18]. Notably, mouse models harboring intestinal knockout of GRP94 led to WNT signaling defects through loss of the WNT co-receptor LRP6, resulting in early postnatal death with loss of intestinal barrier function, decreased number of villi and significant reduction in crypts [50][18].
IGF-1 plays an important role in intestinal growth [51][19] and is a bioactive hormone of HM [52][20]. The IGF system proteins are located in the gastric glands and epithelium and in the apical portion of the villous epithelium of the duodenum. Treatment with porcine MEX promoted IPEC-J2 cell proliferation, raised mice’ villus height, crypt depth and the ratio of villus length to crypt depth of intestinal tissues. MEX also increased CDX2, PCNA and IGF1R and inhibited p53 expression [54][21].
IGF-1 not only promotes growth of the GI tract [51[19][22],53], but protects IECs from oxidative stress, hypoxia, thermal stress and apoptosis in the setting of intestinal injury [55,56,57,58][23][24][25][26]. Moreover, IGF-1 exerts anti-inflammatory properties [59][27], promotes the development and cytotoxic activity of human NK cells [60][28], improves intestinal barrier function [61,62][29][30] and decreases bacterial translocation [63][31]. IGF-1 has thus been suggested to play a promising role in the treatment or prevention of necrotizing enterocolitis (NEC) [51,64,65][19][32][33].
IGF1gene expression is induced byIGF1P2 promoter demethylation [66,67,68][34][35][36]. It has been demonstrated that DNMT1 silencing significantly increases the expression of IGF-1, whereas DNMT1 up-regulation directly results in hypermethylation ofIGF1,thereby reducing IGF-1 expression [69][37]. In accordance, MEX derived from bovine [70][38], porcine [54][21], rat [71][39] and yak milk [72,73][40][41] promote proliferation and survival of IECs. Notably, metabolic activity of human colorectal adenocarcinoma epithelial (Caco-2) cells after co-incubation with bovine colostrum and MEX from high immune responder cows was significantly greater than after co-incubation with MEX from low immune responder cows pointing towards immune-genetic variations of MEX bioactivity [74][42].
Leucine-rich-repeat-containing G-protein-coupled receptor 5 (LGR5), a WNT target gene with restricted crypt expression, has been identified as marker for intestinal stem cells (ISCs) [75][43]. Recent evidence indicates that MEX interact with ISCs. Human MEX exposure to H2O2-treated prominin-1+ISCs derived from small intestines of the neonatal rat increased ISC viability compared to MEX-free controls [77][44]. To elucidate the mechanism by which MEX act in promoting cell growth, Hock et al.
The intestinal epithelium establishes a selectively permeable barrier that supports nutrient absorption and waste secretion while preventing intrusion by luminal materials. The appropriate maturation of the intestinal permeability barrier is of critical importance for the neonate and is often immature in preterm infants, who are at increased risk for developing NEC associated with disrupted tight junctions (TJs) [79,80][45][46]. Formation of functional TJs is critical for the maintenance of gut permeability and intestinal barrier function [78,81][47][48]. zonula occludens 1 (ZO-1) are considered crucial for creating the seal and thus regulate intestinal permeability [78,82,83][47][49][50].
Remarkably, bovine MEX derived from the 100,000×gultracentrifugation fraction of commercial cow milk restored the expression of ZO-1, which was diminished by dextran sodium sulfate (DSS) in a DSS-induced murine model of colitis [84][51]. It has recently been demonstrated that porcine MEX attenuated deoxynivalenol (DON)-induced damage of IECs. OCLN mRNA and protein in IPEC-J2 cells and the small intestinal tissues during continuous DON exposures could be significantly rescued by porcine MEX [85][52]. In accordance, human MEX administration 6 h prior to induction of experimental NEC, showed milder intestinal tissue injury than controls and had lower levels of pro-inflammatory cytokines and higher levels of epithelial TJ proteins ZO-1, claudin and occludin [86][53].
The IECs are covered by a thick layer of mucus, which is produced by goblet cells. Secreted/gel-forming mucins such as MUC2 are responsible for the formation of the mucus layer over the epithelium, whereas the transmembrane mucins such as MUC1 are poorly understood [87,88][54][55]. Mucus in the small intestine forms a diffusion barrier where anti-microbial substances keep the epithelium free from microorganisms [90][56]. Thus, goblet cells and their secreted mucins play a critical role in intestinal barrier function and immune homeostasis.
[47][57] investigated the effects of bovine MEX on goblet cell expression in experimental NEC. To study the effect on mucin production, human colonic LS174T cells were cultured and exposed to bovine MEX. Compared to the control, bovine MEX promoted goblet cell activity, as demonstrated by increased mucin production and relative expression levels of goblet cell expression markers trefoil factor 3 (TFF3) and MUC2 [47][57]. Quantification of immunostaining revealed no difference in goblet cell numbers between raw and HoP human MEX.
Among the factors influencing the mucus barrier, the microbiome plays a major role in driving mucus changes [88][55]. Mucus forms large pores and is penetrable to bacteria and other components, but despite this, in normal situations, the contact between bacteria and the epithelium is limited [93,94][58][59]. The continuous secretion of mucus and its flow towards the intestinal lumen donates anti-bacterial agents including lysozyme, deleted in malignant brain tumors 1 (DMBT1), immunoglobulin A (IgA), defensins, regenerating islet-derived 3γ (RegIIIγ) and phospholipase A2-IIA, which all keep bacteria away from the epithelial surface [88,95,96,97][55][60][61][62].
mice was coupled to increased bacterial colonization of the intestinal epithelial surface and enhanced activation of intestinal adaptive immune responses by the microbiota [98][63]. Thus, RegIIIγ is a fundamental mechanism of innate immunity that promotes host-bacterial mutualism by regulating the spatial relationships between microbiota and host [99][64]. RegIIIγ expression depends on MyD88-mediated signaling downstream of toll-like receptors and the IL-1 receptor family, which is critically involved in the induction of protective host responses upon infections [100][65]. Functional expression of MyD88 in IECs protected mice during intestinal infection, which was associated with enhanced epithelial barrier integrity and increased expression of the RegIIIγ [100][65].
In fact, bovine MEX have been shown to alter bacterial gene expression promoting the growth ofEscherichia coliK-12 Notably, lachnospiraceae, which are butyrate-producing intestinal bacteria [105[66][67][68],106,107], exhibit reduced abundance in ulcerative colitis [108][69]. Remarkably, children with lower risk of IgE-mediated allergic diseases showed an earlier maturation of gut microbiota and an increased abundance of butyrate-producing bacteria, associated with earlier maturation of regulatory T (Treg) cells and lower IgE production [109][70]. The increase in highly activated Treg cells was associated with a relative abundance ofBifidobacterium longumfollowed by increased colonization with butyrate-producing bacteria [109][70].
Intestinal Treg cells are crucial to maintain immune tolerance to dietary antigens and gut microbiota [111][71]. The differentiation, migration and maintenance of intestinal Treg (iTreg) cells are controlled by specific signals from the local environment [114][72]. Intestinal tolerance requires gut homing and expansion of FOXP3+Treg cells in the lamina propria [115][73]. Antigen can be acquired directly by intestinal phagocytes, or pass through enterocytes or goblet cell-associated passages prior to capture by DCs in the lamina propria.
Importantly, DNA demethylation regulates stable FOXP3 expression associated with selective demethylation of an evolutionarily conserved element within theFOXP3locus named TSDR (Treg-specific demethylated region) [117,118,119,120,121][74][75][76][77][78]. In CD4+T cells, the DNA methyltransferases DNMT1 and DNMT3b reside within theFOXP3locus and function to methylate CpG residues, thereby repressing FOXP3 expression in CD4+cells, whereas complete demethylation of this site is required for stable FOXP3 expression [122][79]. Epigenetic regulation ofFOXP3can be predictably controlled with DNMT inhibitors to generate functional, stable and specific Treg cells [123][80].
Exosomes play a pivotal role in important aspects of immune regulation and signaling between various cells of the immune system [126[81][82],127], especially in inflammatory bowel diseases [128,129][83][84]. [130][85] observed increased numbers of FOXP3+CD4+CD25+Treg cells in peripheral blood mononuclear cells (PBMC) incubated with human MEX. In accordance, rat pups exposed to β-lactoglobulin (BLG), one of the main allergenic proteins in cow milk, in the presence of maternal rat milk developed an immune response profile similar to that of unchallenged dam-reared rats associated with a greater FOXP3 expression and increased numbers of FOXP3+CD4+T cells [131][86]. Treg cells via exosome release transfer miRNAs, including let-7d, let-7b and miRNA-155, to conventional T cells.
Remarkably, increased Treg cell numbers are associated with raw farm milk exposure and lower atopic sensitization and asthma in childhood [136][87]. Of note, the protective effect of farm milk consumption on childhood asthma and atopy was lost when boiled farm milk was consumed instead of raw cow milk, pointing to a heat-labile protective factor in milk [137,138][88][89]. There is evidence that vigorous heat-treatment such as ultraheat-treatment (UHT: 135 °C, > 1 s) and boiling (100 °C) of commercial cow milk destroys MEVs and MEX and their miRNA cargo, including miRNA-148a [139[90][91],140], whereas pasteurization (72–78 °C, >15 s) of commercial milk did not affect total MEV numbers and preserved nearly 25–40% of milk´s total small RNAs, including miRNA-148a [139][90]. In comparison to high pressure processing of HM, HoP of HM (62.5 °C, 30 min) resulted in a significant decrease in MEX numbers [22][92].
Thus, early-life exposure to unpasteurized milk may protect against atopy, asthma and related conditions, independently of the place of residence and farming status, in both children and adults [141][93]. HM and unprocessed farm milk may enhance DNMT1-dependent stable Treg cell maturation. A recent randomized controlled trial showed that preterm neonates who received bovine colostrum had higher FOXP3 Treg cell levels compared to controls [142][94].
Interleukin 2 (IL-2) and transforming growth factor-β1 (TGF-β1) also play a central role in Treg cell homeostasis. Strong TCR signal inactivates glycogen synthase kinase 3 (GSK3) to rescue DNMT1 protein from proteasomal degradation and suppresses miRNA-148a to derepress Interestingly, commercial cow milk contains MEX expressing immune-regulatory TGF-β [147][95]. Of note, TGF-β1 was significantly less secreted into mature milk of allergic mothers compared to non-allergic mothers [151][96].
miRNA-155 is another miRNA necessary for the development of Treg cells [152,153][97][98]. Notably, miRNA-155 is highly expressed in human and bovine milk [154,155][99][100]. via targeting signal transducer and activator of transcription 1 (SOCS1) may activate IL-2/STAT5 signaling which promotes Treg cell development [156,157][101][102]. Both FOXP3 and TGF-β increase the expression of miRNA-155 [152,158[97][103][104],159], which plays a key role in the activation and differentiation of iTreg and thymic Treg (tTreg) cells [152,153][97][98].
Recent evidence indicates that the expression of uncoupling protein 3 (UCP3) is involved in the regulation of Treg cells [160][105]. When compared to UCP3+/+mice, CD4+T cells from UCP3-/-mice had increased FOXP3 expression under iTreg cell conditions [160][105]. Notably, UCP3 is a direct target of miRNA-148a. MEX via transfer of miRNA-148a, miRNA-155 and TGF-
It has been demonstrated in various experimental models of NEC that the addition of human, bovine and porcine MEX attenuated the expression of inflammatory cytokines such as interleukin 6 (IL-6), interleukin 1β (IL-1β) and tumor necrosis factor-α (TNF-α) , TLR4 [162,163][106][107] and nuclear factor κB [162][106]. Previous studies showed that miRNA-146a, miRNA-155, miRNA-125b and miRNA-21, abundant immune-regulatory miRNAs of human and bovine milk and MEX [14,22,25,32,164][108][92][10][12][109], inhibit TLR-triggered production of inflammatory cytokines [165,166,167,168,169][110][111][112][113][114].
In phagocytes, changes in cytosolic Ca2+regulate receptor-mediated endocytosis, phagosome-lysosome fusion and antigen processing. There is recent evidence that miRNAs are critically involved in the regulation of DC differentiation and function [171][115]. CaMKII inhibitors blocked the antigen-induced increase in total cellular MHC class molecules as well as their trafficking to the plasma membrane, which was associated with decreased presentation of particulate and soluble MHC class II-restricted antigen [170,173][116][117]. CaMKII has been identified as an activator of IκB kinase (IKK) specifically in response to TCR stimulation [174][118].
Furthermore, miRNA-148a was found to be a direct repressor of IκB kinase β (IKKβ) encoded onIKBKB[176][119]. IKKβ via phosphorylation of IκB results in dissociation of IκB from NF-κB allowing NF-κB translocation to the nucleus, which induces the synthesis of pro-inflammatory cytokines [177][120]. Apparently, MEX-derived miRNA-148a, miRNA-146a, miRNA-155, miRNA-125b and miRNA-21 in a synergistic fashion negatively regulate the activation of immune cells and prevent over-activation of immune responses. signaling may play a key role in the regulation of immune homeostasis and intestinal inflammation [178][121].
[124,125][122][123], physically associates with the Rel family transcription factors, nuclear factor of activated T cells (NFAT) and NF-κB, and blocks their ability to induce the endogenous expression of key pro-inflammatory cytokine genes [179,180][124][125]. Thus, miRNA-148a, the most abundant miRNA of HM and MEX [22][92], interrupts NF-κB signaling at multiple immune-regulatory checkpoints: CaMKII, IKKβ and FOXP3 (Figure 3). signaling relies on the induction of IκB, which traps activated NF-κB in inactive cytoplasmic complexes [181,182,183][126][127][128] , MEX-derived miRNA-148a operates in a synergistic fashion with corticosteroids maintaining high cellular levels of IκB that attenuates pro-inflammatory NF-κB signaling.
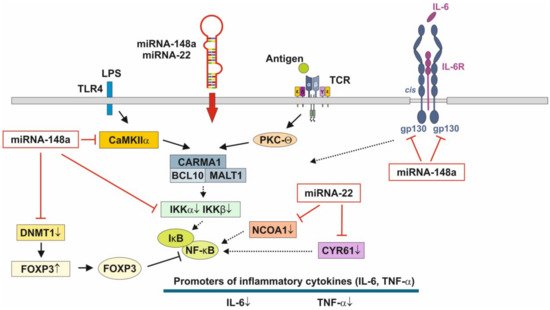
The pathway with the most significant enrichment in miRNA targets from preterm HM is glycosphingolipid biosynthesis [184][129], which is important for neurodevelopment, membrane function and signal transduction of lipid rafts [184][129]. [15][130] demonstrated significant differences in MEX miRNA composition between the HM of mothers delivering preterm infants compared to the HM produced for term infants. The abundant miRNAs in preterm MEX are similar to those from term MEX, whereas 21 low abundance miRNAs are specifically expressed in preterm MEX compared to early term MEX [15][130]. Notably, miRNA-22 is highly expressed in extremely preterm MEX followed by miRNA-148a [15][130].
Furthermore, miRNA-22 is involved in the regulation of metabolism, energy expenditure and immune functions. Important targets of miRNA-22 are PTEN, purine rich element binding protein B (PURB), caveolin 3 (CAV3), -γ co-activator 1α (PGC-1α), peroxisome proliferator-activated receptor-α (PPARα) and sirtuin 1 (SIRT1), which coordinate fatty acid metabolism, mitochondrial biogenesis and energy homeostasis [186,187][131][132]. Loss of miRNA-22 reduces fat mass gain induced by high-fat diet and enhanced energy expenditure [186,188][131][133].
miRNA-22 exerts strong anti-inflammatory activities via targeting the mRNA of cysteine-rich protein 61 [189][134], a component of the extracellular matrix, which is produced and secreted by several cell types including endothelial cells, fibroblasts and smooth muscle cells. CYR61 was mainly up-regulated in intestinal mucosa after intestinal ischemia/reperfusion injury in pigs [192][135]. In addition, miRNA-22 attenuates myocardial ischemia-reperfusion injury via an anti-inflammatory mechanism in rats [193][136] and via targeting CREB binding protein (CBP) protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats [194][137].
It has been demonstrated in murine macrophages that CYR61 activates NF-κB-mediated transcription, and induces a pro-inflammatory genetic program characteristic of classically activated M1 macrophages that participates in Th1 responses. miRNA-22 over- expression significantly inhibited NF-κB activity by decreasing nuclear receptor co-activator 1 (NCOA1) expression (Figure 3) [196,197][138][139]. Thus, over-expressed miRNA-22 in MEX delivered to preterm infants with low birthweight appear to promote growth, weight gain, tissue maturation and attenuates inflammatory responses. This suggests that preterm milk and their MEX-derived miRNAs may have adaptive functions for growth and maturation in premature infants.
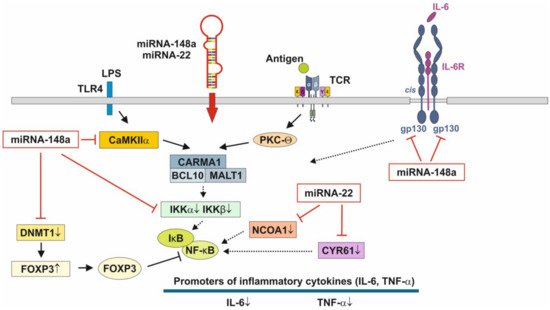
Figure 3. Anti-inflammatory actions of miRNA-148a and miRNA-22 on nuclear factor κB signaling. miRNA-148a via suppression of DNA methyltransferase 1 (DNMT1) enhances the expression of FOXP3, which is a negative regulator of nuclear factor κB. miRNA-148a directly targets calcium/calmodulin-dependent protein kinase IIα (CaMKIIα), which phosphorylates CARD-containing MAGUK protein 1 (CARMA1) involved in the activation of IκB kinase α (IKKα) and IκB kinase β (IKKβ), Notably, miRNA-148a directly targets IKKα and IKKβ, thereby enhancing the inhibitory effect of IκB on NF-κB. In addition, miRNA-148a targets the interleukin 6 (IL-6) signal transducer gp130. miRNA-22, which is highly expressed in preterm MEX, targets nuclear receptor co-activator 1 (NCOA1) and cystein-rich protein 61 (CYR61), which both activate NF-κB. miRNA-30b via targeting RIP140 suppresses IL-6 expression. MEX-derived miRNAs thus provide anti-inflammatory signaling.
Murine and human Lgr5+ISCs showed high expression of the immune cell-associated circRNA circPan3 [202][140]. circRNAs are related to inflammatory bowel disease and intestinal barrier formation [203][141]. [199][142] identified 6756 circRNAs both in preterm human colostrum and term colostrum, of which 66 were up-regulated and 42 were down-regulated in preterm colostrum. In particular, MEX found in preterm colostrum and term colostrum promoted VEGF protein expression and induced the proliferation and migration of small IECs [199][142].
Taken together, the physiological adaptations of colostrum and MEX RNAs in the milk of mothers, who delivered preterm infants, may accelerate intestinal maturation, barrier function and innate immunity, critical factors for the prevention of NEC.
3. Milk Processing and Exosome Bioavailability
There is recent interest to use MEX and their miRNA cargo for the treatment and prevention of NEC [230][143] and to supplement MEX-deficient artificial formula [460][144]. Whereas UHT (135 °C, >1 s) and boiling (100 °C) of commercial cow milk destroys MEVs and MEX and their miRNA cargo [139,140][90][91], pasteurization (72–78 °C, >15 s) of commercial cow milk did not affect MEV numbers and preserved nearly 25–40% of milk’s total small RNAs [139][90]. However, the effects of UV-C irradiation on MEX structure and bioavailbilty have not yet been studied. Notably, lyophilization of exosomes without the cryoprotectant trehalose results in exosome aggregation, while the addition of trehalose prevents aggregation during lyophilization [466,467][145][146].
References
- Renz, H.; Brandtzaeg, P.; Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011, 12, 9–23.
- Zonneveld, M.I.; van Herwijnen, M.J.C.; Fernandez-Gutierrez, M.M.; Giovanazzi, A.; de Groot, A.M.; Kleinjan, M.; van Capel, T.M.M.; Sijts, A.J.A.M.; Taams, L.S.; Garssen, J.; et al. Human milk extracellular vesicles target nodes in interconnected signalling pathways that enhance oral epithelial barrier function and dampen immune responses. J. Extracell. Vesicles 2021, 10, e12071.
- Carr, L.E.; Virmani, M.D.; Rosa, F.; Munblit, D.; Matazel, K.S.; Elolimy, A.A.; Yeruva, L. Role of human milk bioactives on infants’ gut and immune health. Front. Immunol. 2021, 12, 604080.
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490.
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103.
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087.
- Melnik, B.C.; Kakulas, F.; Geddes, D.T.; Hartmann, P.E.; John, S.M.; Carrera-Bastos, P.; Cordain, L.; Schmitz, G. Milk miRNAs: Simple nutrients or systemic functional regulators? Nutr. Metab. 2016, 13, 42.
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564.
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009.
- Van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.; Wauben, M.H.M. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front. Nutr. 2018, 5, 81.
- Chen, Z.; Xie, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. Milk exosome-derived miRNAs from water buffalo are implicated in immune response and metabolism process. BMC Vet. Res. 2020, 16, 123.
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123.
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641.
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol. Neurobiol. 2016, 36, 301–312.
- Jadli, A.S.; Ballasy, N.; Edalat, P.; Patel, V.B. Inside(sight) of tiny communicator: Exosome biogenesis, secretion, and uptake. Mol. Cell Biochem. 2020, 467, 77–94.
- Wolf, T.; Baier, S.R.; Zempleni, J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 Cells. J. Nutr. 2015, 145, 2201–2206.
- Reif, S.; Elbaum Shiff, Y.; Golan-Gerstl, R. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J. Transl. Med. 2019, 17, 325.
- Liu, B.; Staron, M.; Hong, F.; Wu, B.X.; Sun, S.; Morales, C.; Crosson, C.E.; Tomlinson, S.; Kim, I.; Wu, D.; et al. Essential roles of grp94 in gut homeostasis via chaperoning canonical Wnt pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 6877–6882.
- Shelby, R.D.; Cromeens, B.; Rager, T.M.; Besner, G.E. Influence of growth factors on the development of necrotizing enterocolitis. Clin. Perinatol. 2019, 46, 51–64.
- Hoeflich, A.; Meyer, Z. Functional analysis of the IGF-system in milk. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 409–418.
- Chen, T.; Xie, M.Y.; Sun, J.J.; Ye, R.S.; Cheng, X.; Sun, R.P.; Wei, L.M.; Li, M.; Lin, D.L.; Jiang, Q.Y.; et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci. Rep. 2016, 6, 33862.
- Freier, S.; Eran, M.; Reinus, C.; Ariel, I.; Faber, J.; Wilschanski, M.; Braverman, D. Relative expression and localization of the insulin-like growth factor system components in the fetal, child and adult intestine. J. Pediatr. Gastroenterol. Nutr. 2005, 40, 202–209.
- Ozen, S.; Akisu, M.; Baka, M.; Yalaz, M.; Sozmen, E.Y.; Berdeli, A.; Kultursay, N. Insulin-like growth factor attenuates apoptosis and mucosal damage in hypoxia/reoxygenation-induced intestinal injury. Biol. Neonate 2005, 87, 91–96.
- Wilkins, H.R.; Ohneda, K.; Keku, T.O.; D’Ercole, A.J.; Fuller, C.R.; Williams, K.L.; Lund, P.K. Reduction of spontaneous and irradiation-induced apoptosis in small intestine of IGF-I transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, G457–G464.
- Jeschke, M.G.; Bolder, U.; Chung, D.H.; Przkora, R.; Mueller, U.; Thompson, J.C.; Wolf, S.E.; Herndon, D.N. Gut mucosal homeostasis and cellular mediators after severe thermal trauma and the effect of insulin-like growth factor-I in combination with insulin-like growth factor binding protein-3. Endocrinology 2007, 148, 354–362.
- Baregamian, N.; Rychahou, P.G.; Hawkins, H.K.; Evers, B.M.; Chung, D.H. Phosphatidylinositol 3-kinase pathway regulates hypoxia-inducible factor-1 to protect from intestinal injury during necrotizing enterocolitis. Surgery 2007, 142, 295–302.
- Tian, F.; Liu, G.R.; Li, N.; Yuan, G. Insulin-like growth factor I reduces the occurrence of necrotizing enterocolitis by reducing inflammatory response and protecting intestinal mucosal barrier in neonatal rats model. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4711–4719.
- Ni, F.; Sun, R.; Fu, B.; Wang, F.; Guo, C.; Tian, Z.; Wei, H. IGF-1 promotes the development and cytotoxic activity of human NK cells. Nat. Commun. 2013, 4, 1479.
- Zhang, W.; Frankel, W.L.; Adamson, W.T.; Roth, J.A.; Mantell, M.P.; Bain, A.; Ziegler, T.R.; Smith, R.J.; Rombeau, J.L. Insulin-like growth factor-I improves mucosal structure and function in transplanted rat small intestine. Transplantation 1995, 59, 755–761.
- Lorenzo-Zúñiga, V.; Rodríguez-Ortigosa, C.M.; Bartolí, R.; Martínez-Chantar, M.L.; Martínez-Peralta, L.; Pardo, A.; Ojanguren, I.; Quiroga, J.; Planas, R.; Prieto, J. Insulin-like growth factor I improves intestinal barrier function in cirrhotic rats. Gut 2006, 55, 1306–1312.
- Hunninghake, G.W.; Doerschug, K.C.; Nymon, A.B.; Schmidt, G.A.; Meyerholz, D.K.; Ashare, A. Insulin-like growth factor-1 levels contribute to the development of bacterial translocation in sepsis. Am. J. Respir. Crit. Care Med. 2010, 182, 517–525.
- Corpeleijn, W.E.; van Vliet, I.; de Gast-Bakker, D.A.; van der Schoor, S.R.; Alles, M.S.; Hoijer, M.; Tibboel, D.; van Goudoever, J.B. Effect of enteral IGF-1 supplementation on feeding tolerance, growth, and gut permeability in enterally fed premature neonates. J. Pediatr. Gastroenterol. Nutr. 2008, 46, 184–190.
- Rowland, K.J.; Choi, P.M.; Warner, B.W. The role of growth factors in intestinal regeneration and repair in necrotizing enterocolitis. Semin. Pediatr. Surg. 2013, 22, 101–111.
- Ouni, M.; Gunes, Y.; Belot, M.P.; Castell, A.L.; Fradin, D.; Bougnères, P. The IGF1 P2 promoter is an epigenetic QTL for circulating IGF1 and human growth. Clin. Epigenetics 2015, 7, 22.
- Ouni, M.; Castell, A.L.; Linglart, A.; Bougnères, P. Genetic and epigenetic modulation of growth hormone sensitivity studied with the IGF-1 generation test. J. Clin. Endocrinol. Metab. 2015, 100, E919–E925.
- Ouni, M.; Belot, M.P.; Castell, A.L.; Fradin, D.; Bougnères, P. The P2 promoter of the IGF1 gene is a major epigenetic locus for GH responsiveness. Pharmacogenomics J. 2016, 16, 102–106.
- Ma, M.; Zhou, Q.J.; Xiong, Y.; Li, B.; Li, X.T. Preeclampsia is associated with hypermethylation of IGF-1 promoter mediated by DNMT1. Am. J. Transl. Res. 2018, 10, 16–39.
- Yu, S.; Zhao, Z.; Sun, L.; Li, P. Fermentation results in quantitative changes in milk-derived exosomes and different effects on cell growth and survival. J. Agric. Food Chem. 2017, 65, 1220–1228.
- Hock, A.; Miyake, H.; Li, B.; Lee, C.; Ermini, L.; Koike, Y.; Chen, Y.; Määttänen, P.; Zani, A.; Pierro, A. Breast milk-derived exosomes promote intestinal epithelial cell growth. J. Pediatr. Surg. 2017, 52, 755–759.
- Gao, H.N.; Guo, H.Y.; Zhang, H.; Xie, X.L.; Wen, P.C.; Ren, F.Z. Yak-milk-derived exosomes promote proliferation of intestinal epithelial cells in an hypoxic environment. J. Dairy Sci. 2019, 102, 985–996.
- Gao, H.N.; Ren, F.Z.; Wen, P.C.; Xie, L.X.; Wang, R.; Yang, Z.N.; Li, Y.X. Yak milk-derived exosomal microRNAs regulate intestinal epithelial cells on proliferation in hypoxic environment. J. Dairy Sci. 2021, 104, 1291–1303.
- Ross, M.; Atalla, H.; Karrow, N.; Mallard, B.A. The bioactivity of colostrum and milk exosomes of high, average, and low immune responder cows on human intestinal epithelial cells. J. Dairy Sci. 2021, 104, 2499–2510.
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007.
- Dong, P.; Zhang, Y.; Yan, D.Y.; Wang, Y.; Xu, X.; Zhao, Y.C.; Xiao, T.T. Protective effects of human milk-derived exosomes on intestinal stem cells damaged by oxidative stress. Cell Transplant. 2020, 29, 963689720912690.
- Ravisankar, S.; Tatum, R.; Garg, P.M.; Herco, M.; Shekhawat, P.S.; Chen, Y.H. Necrotizing enterocolitis leads to disruption of tight junctions and increase in gut permeability in a mouse model. BMC Pediatr. 2018, 18, 372.
- Liu, D.; Xu, Y.; Feng, J.; Yu, J.; Huang, J.; Li, Z. Mucins and tight junctions are severely altered in necrotizing enterocolitis neonates. Am. J. Perinatol. 2020.
- Buckley, A.; Turner, J.R. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a029314.
- Camilleri, M.; Madsen, K.; Spiller, R.; Greenwood-Van Meerveld, B.; Verne, G.N. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol. Motil. 2012, 24, 503–512.
- Furuse, M.; Hata, M.; Furuse, K.; Yoshida, Y.; Haratake, A.; Sugitani, Y.; Noda, T.; Kubo, A.; Tsukita, S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 2002, 156, 1099–1111.
- Fanning, A.S.; Jameson, B.J.; Jesaitis, L.A.; Anderson, J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 1998, 273, 29745–29753.
- Benmoussa, A.; Diallo, I.; Salem, M.; Michel, S.; Gilbert, C.; Sévigny, J.; Provost, P. Concentrates of two subsets of extracellular vesicles from cow’s milk modulate symptoms and inflammation in experimental colitis. Sci. Rep. 2019, 9, 14661.
- Xie, M.Y.; Chen, T.; Xi, Q.Y.; Hou, L.J.; Luo, J.Y.; Zeng, B.; Li, M.; Sun, J.J.; Zhang, L. Porcine milk exosome miRNAs protect intestinal epithelial cells against deoxynivalenol-induced damage. Biochem. Pharmacol. 2020, 175, 113898.
- He, S.; Liu, G.; Zhu, X. Human breast milk-derived exosomes may help maintain intestinal epithelial barrier integrity. Pediatr. Res. 2021.
- Cornick, S.; Tawiah, A.; Chadee, K. Roles and regulation of the mucus barrier in the gut. Tissue Barriers 2015, 3, e982426.
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243.
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649.
- Li, B.; Hock, A.; Wu, R.Y.; Minich, A.; Botts, S.R.; Lee, C.; Antounians, L.; Miyake, H.; Koike, Y.; Chen, Y.; et al. Bovine milk-derived exosomes enhance goblet cell activity and prevent the development of experimental necrotizing enterocolitis. PLoS ONE 2019, 14, e0211431.
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526.
- Hansson, G.C. Mucins and the microbiome. Annu. Rev. Biochem. 2020, 89, 769–793.
- Johansson, M.E.V.; Sjövall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361.
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 2014, 260, 8–20.
- Schroeder, B.O. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019, 7, 3–12.
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334, 255–258.
- Santaolalla, R.; Abreu, M.T. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 2012, 28, 124–129.
- Friedrich, C.; Mamareli, P.; Thiemann, S.; Kruse, F.; Wang, Z.; Holzmann, B.; Strowig, T.; Sparwasser, T.; Lochner, M. MyD88 signaling in dendritic cells and the intestinal epithelium controls immunity against intestinal infection with C. rodentium. PLoS Pathog. 2017, 13, e1006357.
- Zhang, J.; Song, L.; Wang, Y.; Liu, C.; Zhang, L.; Zhu, S.; Liu, S.; Duan, L. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol. 2019, 34, 1368–1376.
- Sasaki, K.; Inoue, J.; Sasaki, D.; Hoshi, N.; Shirai, T.; Fukuda, I.; Azuma, T.; Kondo, A.; Osawa, R. Construction of a model culture system of human colonic microbiota to detect decreased Lachnospiraceae abundance and butyrogenesis in the feces of ulcerative colitis patients. Biotechnol. J. 2019, 14, e1800555.
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455.
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450.
- Ruohtula, T.; de Goffau, M.C.; Nieminen, J.K.; Honkanen, J.; Siljander, H.; Hämäläinen, A.M.; Peet, A.; Tillmann, V.; Ilonen, J.; Niemelä, O.; et al. Maturation of gut microbiota and circulating regulatory T cells and development of IgE sensitization in early life. Front. Immunol. 2019, 10, 2494.
- Yamashiro, Y. Gut microbiota in health and disease. Ann. Nutr. Metab. 2017, 71, 242–246.
- Tanoue, T.; Atarashi, K.; Honda, K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 2016, 16, 295–309.
- Hadis, U.; Wahl, B.; Schulz, O.; Hardtke-Wolenski, M.; Schippers, A.; Wagner, N.; Müller, W.; Sparwasser, T.; Förster, R.; Pabst, O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 2011, 34, 237–246.
- Huehn, J.; Beyer, M. Epigenetic and transcriptional control of Foxp3+ regulatory T cells. Semin. Immunol. 2015, 27, 10–18.
- Bellanti, J.A.; Li, D. Treg cells and epigenetic regulation. Adv. Exp. Med. Biol. 2021, 1278, 95–114.
- Polansky, J.K.; Kretschmer, K.; Freyer, J.; Floess, S.; Garbe, A.; Baron, U.; Olek, S.; Hamann, A.; von Boehmer, H.; Huehn, J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008, 38, 1654–1663.
- Toker, A.; Engelbert, D.; Garg, G.; Polansky, J.K.; Floess, S.; Miyao, T.; Baron, U.; Düber, S.; Geffers, R.; Giehr, P.; et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 2013, 190, 3180–3188.
- Schreiber, L.; Pietzsch, B.; Floess, S.; Farah, C.; Jänsch, L.; Schmitz, I.; Huehn, J. The Treg-specific demethylated region stabilizes Foxp3 expression independently of NF-κB signaling. PLoS ONE 2014, 9, e88318.
- Lal, G.; Bromberg, J.S. Epigenetic mechanisms of regulation of Foxp3 expression. Blood 2009, 114, 3727–3735.
- Lal, G.; Zhang, N.; van der Touw, W.; Ding, Y.; Ju, W.; Bottinger, E.P.; Reid, S.P.; Levy, D.E.; Bromberg, J.S. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 2009, 182, 259–273.
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208.
- Seo, N. Exosome-mediated immune regulation and its clinical application. Trends Immunother. 2020, 4, 36–41.
- Wani, S.; Man Law, I.K.; Pothoulakis, C. Role and mechanisms of exosomal miRNAs in IBD pathophysiology. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G646–G654.
- Ocansey, D.K.W.; Zhang, L.; Wang, Y.; Yan, Y.; Qian, H.; Zhang, X.; Xu, W.; Mao, F. Exosome-mediated effects and applications in inflammatory bowel disease. Biol. Rev. Camb. Philos. Soc. 2020, 95, 1287–1307.
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978.
- Tooley, K.L.; El-Merhibi, A.; Cummins, A.G.; Grose, R.H.; Lymn, K.A.; DeNichilo, M.; Penttila, I.A. Maternal milk, but not formula, regulates the immune response to beta-lactoglobulin in allergy-prone rat pups. J. Nutr. 2009, 139, 2145–2151.
- Lluis, A.; Depner, M.; Gaugler, B.; Saas, P.; Casaca, V.I.; Raedler, D.; Michel, S.; Tost, J.; Liu, J.; Genuneit, J.; et al. Increased regulatory T-cell numbers are associated with farm milk exposure and lower atopic sensitization and asthma in childhood. J. Allergy Clin. Immunol. 2014, 133, 551–559.
- Loss, G.; Apprich, S.; Waser, M.; Kneifel, W.; Genuneit, J.; Büchele, G.; Weber, J.; Sozanska, B.; Danielewicz, H.; Horak, E.; et al. The protective effect of farm milk consumption on childhood asthma and atopy: The GABRIELA study. J. Allergy Clin. Immunol. 2011, 128, 766–773.
- Brick, T.; Hettinga, K.; Kirchner, B.; Pfaffl, M.W.; Ege, M.J. The beneficial effect of farm milk consumption on asthma, allergies, and infections: From meta-analysis of evidence to clinical trial. J. Allergy Clin. Immunol. Pract. 2020, 8, 878–889.
- Kirchner, B.; Pfaffl, M.W.; Dumpler, J.; von Mutius, E.; Ege, M.J. microRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 2016, 137, 1893–1895.
- Kleinjan, M.; van Herwijnen, M.J.; Libregts, S.F.; van Neerven, R.J.; Feitsma, A.L.; Wauben, M.H. Regular industrial processing of bovine milk impacts the integrity and molecular composition of extracellular vesicles. J. Nutr. 2021.
- Smyczynska, U.; Bartlomiejczyk, M.A.; Stanczak, M.M.; Sztromwasser, P.; Wesolowska, A.; Barbarska, O.; Pawlikowska, E.; Fendler, W. Impact of processing method on donated human breast milk microRNA content. PLoS ONE 2020, 15, e0236126.
- Sozańska, B.; Pearce, N.; Dudek, K.; Cullinan, P. Consumption of unpasteurized milk and its effects on atopy and asthma in children and adult inhabitants in rural Poland. Allergy 2013, 68, 644–650.
- Ismail, R.I.H.; Awad, H.A.; Imam, S.S.; Gad, G.I.; Aboushady, N.M.; Abdou, R.M.; Eissa, D.S.; Azzam, N.T.; Barakat, M.M.; Yassin, M.M.; et al. Gut priming with bovine colostrum and T regulatory cells in preterm neonates: A randomized controlled trial. Pediatr. Res. 2021.
- Pieters, B.C.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.; van Caam, A.P.; Koenders, M.I.; van Lent, P.L.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M.; et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123.
- Rigotti, E.; Piacentini, G.L.; Ress, M.; Pigozzi, R.; Boner, A.L.; Peroni, D.G. Transforming growth factor-beta and interleukin-10 in breast milk and development of atopic diseases in infants. Clin. Exp. Allergy 2006, 36, 614–618.
- Kohlhaas, S.; Garden, O.A.; Scudamore, C.; Turner, M.; Okkenhaug, K.; Vigorito, E. Cutting edge: The Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 2009, 182, 2578–2582.
- Yao, R.; Ma, Y.L.; Liang, W.; Li, H.H.; Ma, Z.J.; Yu, X.; Liao, Y.H. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 2012, 7, e46082.
- Na, R.S.; E, G.X.; Sun, W.; Sun, X.W.; Qiu, X.Y.; Chen, L.P.; Huang, Y.F. Expressional analysis of immune-related miRNAs in breast milk. Genet. Mol. Res. 2015, 14, 11371–11376.
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841.
- Lu, L.F.; Thai, T.H.; Calado, D.P.; Chaudhry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewsky, K.; et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009, 30, 80–91.
- Zeng, Q.; Liu, W.; Luo, R.; Lu, G. MicroRNA-181a and microRNA-155 are involved in the regulation of the differentiation and function of regulatory T cells in allergic rhinitis children. Pediatr. Allergy Immunol. 2019, 30, 434–442.
- Li, D.P.; Fan, J.; Wu, Y.J.; Xie, Y.F.; Zha, J.M.; Zhou, X.M. MiR-155 up-regulated by TGF-β promotes epithelial-mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am. J. Transl. Res. 2017, 9, 2956–2965.
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12.
- O’Connor, E.B.; Muñoz-Wolf, N.; Leon, G.; Lavelle, E.C.; Mills, K.H.G.; Walsh, P.T.; Porter, R.K. UCP3 reciprocally controls CD4+ Th17 and Treg cell differentiation. PLoS ONE 2020, 15, e0239713.
- Xie, M.Y.; Hou, L.J.; Sun, J.J.; Zeng, B.; Xi, Q.Y.; Luo, J.Y.; Chen, T.; Zhang, Y.L. Porcine milk exosome miRNAs attenuate LPS-induced apoptosis through inhibiting TLR4/NF-κB and p53 pathways in intestinal epithelial cells. J. Agric. Food Chem. 2019, 67, 9477–9491.
- Gao, R.; Zhang, R.; Qian, T.; Peng, X.; He, W.; Zheng, S.; Cao, Y.; Pierro, A.; Shen, C. A comparison of exosomes derived from different periods breast milk on protecting against intestinal organoid injury. Pediatr. Surg. Int. 2019, 35, 1363–1368.
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082.
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137.
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486.
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158.
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609.
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089.
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147.
- Turner, M.L.; Schnorfeil, F.M.; Brocker, T. MicroRNAs regulate dendritic cell differentiation and function. J. Immunol. 2011, 187, 3911–3917.
- Herrmann, T.L.; Morita, C.T.; Lee, K.; Kusner, D.J. Calmodulin kinase II regulates the maturation and antigen presentation of human dendritic cells. J. Leukoc. Biol. 2005, 78, 1397–1407.
- Herrmann, T.L.; Agrawal, R.S.; Connolly, S.F.; McCaffrey, R.L.; Schlomann, J.; Kusner, D.J. MHC Class II levels and intracellular localization in human dendritic cells are regulated by calmodulin kinase II. J. Leukoc. Biol. 2007, 82, 686–699.
- Hughes, K.; Edin, S.; Antonsson, A.; Grundström, T. Calmodulin-dependent kinase II mediates T cell receptor/CD3- and phorbol ester-induced activation of IkappaB kinase. J. Biol. Chem. 2001, 276, 36008–36013.
- Patel, V.; Carrion, K.; Hollands, A.; Hinton, A.; Gallegos, T.; Dyo, J.; Sasik, R.; Leire, E.; Hardiman, G.; Mohamed, S.A.; et al. The stretch responsive microRNA miR-148a-3p is a novel repressor of IKBKB, NF-κB signaling, and inflammatory gene expression in human aortic valve cells. FASEB J. 2015, 29, 1859–1868.
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651.
- Wullaert, A.; Bonnet, M.C.; Pasparakis, M. NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res. 2011, 21, 146–158.
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk: An exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J. Transl. Med. 2014, 12, 43.
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. Milk: A postnatal imprinting system stabilizing FoxP3 expression and regulatory T cell differentiation. Clin. Transl. Allergy 2016, 6, 18.
- Bettelli, E.; Dastrange, M.; Oukka, M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5138–5143.
- Kim, C.H. FOXP3 and its role in the immune system. Adv. Exp. Med. Biol. 2009, 665, 17–29.
- Auphan, N.; DiDonato, J.A.; Rosette, C.; Helmberg, A.; Karin, M. Immunosuppression by glucocorticoids: Inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science 1995, 270, 286–290.
- Scheinman, R.I.; Cogswell, P.C.; Lofquist, A.K.; Baldwin, A.S., Jr. Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science 1995, 270, 283–286.
- Almawi, W.Y.; Melemedjian, O.K. Negative regulation of nuclear factor-kappaB activation and function by glucocorticoids. J. Mol. Endocrinol. 2002, 28, 69–78.
- Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Silveyra, P.; Podany, A.; Birch, L.L.; Paul, I.M.; Kelleher, S.; Hicks, S.D. Metabolism-related microRNAs in maternal breast milk are influenced by premature delivery. Pediatr. Res. 2017, 82, 226–236.
- Kahn, S.; Liao, Y.; Du, X.; Xu, W.; Li, J.; Lönnerdal, B. Exosomal microRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2018, 62, e1701050.
- Diniz, G.P.; Huang, Z.P.; Liu, J.; Chen, J.; Ding, J.; Fonseca, R.I.; Barreto-Chaves, M.L.; Donato, J., Jr.; Hu, X.; Wang, D.Z. Loss of microRNA-22 prevents high-fat diet induced dyslipidemia and increases energy expenditure without affecting cardiac hypertrophy. Clin. Sci. 2017, 131, 2885–2900.
- Yang, Z.; Qin, W.; Huo, J.; Zhuo, Q.; Wang, J.; Wang, L. MiR-22 modulates the expression of lipogenesis-related genes and promotes hepatic steatosis in vitro. FEBS Open Bio 2021, 11, 322–332.
- Thibonnier, M.; Esau, C.; Ghosh, S.; Wargent, E.; Stocker, C. Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care 2020, 8, e001478.
- Lin, J.; Huo, R.; Xiao, L.; Zhu, X.; Xie, J.; Sun, S.; He, Y.; Zhang, J.; Sun, Y.; Zhou, Z.; et al. A novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 49–59.
- Shegarfi, H.; Krohn, C.D.; Gundersen, Y.; Kjeldsen, S.F.; Hviid, C.V.; Ruud, T.E.; Aasen, A.O. Regulation of CCN1 (Cyr61) in a porcine model of intestinal ischemia/reperfusion. Innate Immun. 2015, 21, 453–462.
- Yang, J.; Fan, Z.; Yang, J.; Ding, J.; Yang, C.; Chen, L. microRNA-22 attenuates myocardial ischemia-reperfusion injury via an anti-inflammatory mechanism in rats. Exp. Ther. Med. 2016, 12, 3249–3255.
- Yang, J.; Chen, L.; Yang, J.; Ding, J.; Li, S.; Wu, H.; Zhang, J.; Fan, Z.; Dong, W.; Li, X. MicroRNA-22 targeting CBP protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats. Mol. Biol. Rep. 2014, 41, 555–561.
- Yu, H.; Wu, M.; Zhao, P.; Huang, Y.; Wang, W.; Yin, W. Neuroprotective effects of viral overexpression of microRNA-22 in rat and cell models of cerebral ischemia-reperfusion injury. J. Cell Biochem. 2015, 116, 233–241.
- Takata, A.; Otsuka, M.; Kojima, K.; Yoshikawa, T.; Kishikawa, T.; Yoshida, H.; Koike, K. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem. Biophys. Res. Commun. 2011, 411, 826–831.
- Zhu, P.; Zhu, X.; Wu, J.; He, L.; Lu, T.; Wang, Y.; Liu, B.; Ye, B.; Sun, L.; Fan, D.; et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat. Immunol. 2019, 20, 183–194.
- Rankin, C.R.; Lokhandwala, Z.A.; Huang, R.; Pekow, J.; Pothoulakis, C.; Padua, D. Linear and circular CDKN2B-AS1 expression is associated with inflammatory bowel disease and participates in intestinal barrier formation. Life Sci. 2019, 231, 116571.
- Zhou, Y.; Yu, Z.; Wang, X.; Chen, W.; Liu, Y.; Zhang, Y.; Yin, J.; Han, S. Exosomal circRNAs contribute to intestinal development via the VEGF signalling pathway in human term and preterm colostrum. Aging 2021.
- Chen, W.; Wang, X.; Yan, X.; Yu, Z.; Zhang, J.; Han, S. The emerging role of exosomes in the pathogenesis, prognosis and treatment of necrotizing enterocolitis. Am. J. Transl. Res. 2020, 12, 7020–7033.
- Zempleni, J.; US Department of Agriculture. Milk Findings May Help Infants Worldwide. Available online: (accessed on 10 May 2021).
- Charoenviriyakul, C.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Preservation of exosomes at room temperature using lyophilization. Int. J. Pharm. 2018, 553, 1–7.
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomed. 2020, 15, 6917–6934.
