In the latest years, fluorescence microscopy-related techniques have exhibited continuous developments and progresses, both from technical and applicative perspectives, holding the promise to provide unprecedented tools for drug delivery research.We have revised the major fluorescence microscopy-related experimental techniques available for the characterization of drug delivery systems (DDS)from static and dynamic points of view in different media, with a particular focus on the investigation within biological environments and in vivo. Indeed, the opportunities provided by fluorescence microscopy-related techniques to disentangle scientific issues typical of drug delivery research (spanning from the colloidal characterization of a DDS to its adhesion to biological membranes, its interaction with biomolecules, and its intracellular behavior) are countless and exponentially growing, allowing for the expectation that in the next few years the development of completely new tools and protocols will truly advance drug delivery research.
- drug delivery
- nanoscopy
- super-resolution fluorescence microscopy
- STED
- STORM
- PALM
- TIRF
- light-sheet microscopy
- FCS
- particle tracking
- FRAP
1. Introduction
The development of nanomaterials for the delivery and controlled release of drugs to a selected, specific biological target has been, for many years, one of the core areas of nanomedicine research. Significant contributions to the accomplishments in the design/synthesis of complex drug delivery systems (DDSs) have resulted from the continuous theoretical progression and the improvements of fundamental knowledge on nanomaterials, as well as from the technical advancements of experimental techniques for nanomaterials characterizations. The gap between the synthesis of the DDSs and their full translation into medical practice is related to poor understandings of their behavior in biological environments [1][2][4,5]. In this respect, many efforts are currently devoted to the improvement of knowledge regarding the fate of nanomaterials designed for biomedical applications in living organisms, and of their behavior within biological fluids and with biologically relevant interfaces, such as cell membranes [2][3][4][5][6][5,6,7,8,9].
In this framework, fluorescence microscopy and laser scanning confocal microscopy (LSCM) are key experimental techniques to unravel the behavior of nanomaterials designed for pharmaceutical applications in biological environments [7][8][9][10][10,11,12,13]. Compared to other imaging techniques, in fluorescence microscopy the signal (and, consequently, the contrast) is provided by fluorescence (or autofluorescence); therefore, choosing the appropriate fluorescent probes allows for the highlighting of specific characteristics of the sample (as hydrophobic/hydrophilic regions), Compared to standard fluorescence microscopy, the confocal setup increases the effective signal-to-noise ratio, thanks to the presence of two pinholes. The advent of confocal microscopy has represented a major advancement in fluorescence microscopy, providing the ability to track the localization of DDSs in complex biological media and to optically reconstruct the three-dimensional space around them with extremely low out-of-focus noise and improved spatial resolution.
In addition, aside from imaging techniques, which provide the ability to unravel the localization of DDSs in complex environments, several fluorescence microscopy-related techniques have been developed and refined over the years, thus allowing for informational gain related to the dynamics of DDSs (i.e., their diffusion modes and rates). Fluorescence recovery after photobleaching (FRAP), fluorescence correlation spectroscopy (FCS), and particle tracking (PT) can provide (at different timescales, on different systems, and with different theoretical frameworks and experimental setups) information on the dynamics of DDSs, or on the modification of the typical dynamics of biological environments in response to the interaction with a DDS. Crucial information on DDS characteristics and behaviors in a complex biological environment can be obtained through these techniques, as the internalization mode of a nanocarrier inside a cell lumen (i.e., the specific internalization route, as well as its internalization form, as an assembled or disassembled entity), its adhesion to a biological interface, and its interaction with relevant biomolecules present within biological fluids [11][12][13][14][14,15,16,17].
Despite the extensive application of LSCM in the characterization of biological systems and nanostructured DDSs, a clear limitation is represented by the achievable spatial resolution, which is far from that of electron microscopes. Indeed, typical sizes of nanostructured DDSs are within the range of a few–a few tens or hundreds of nanometers, which are also the typical length scales of DDS interactions with the surrounding environment, while the resolution limits of LSCM are 200–300 nm in the xy plane and 500–700 nm in the axial direction (depending on the optical setup and on the wavelength of the laser line). Due to this inherent physical limit imposed by the diffraction limit of light, the information obtained through LSCM on the characteristics of nanostructured DDSs, as well as on their impact on subcellular processes or interaction with biological interfaces, is limited.
In recent years, different methods have been developed to break the physical diffraction limit of light, approaching the typical nanometric resolution of electron microscopy. From the recognition of the potential groundbreaking impact of super-resolution imaging, with the 2014 Nobel prize in Chemistry, different nanoscopy techniques have been implemented on commercial microscopes, and are constantly refined to expand their applicability. Driven by this new hype around super-resolution microscopy, a general renewed interest has grown on advanced fluorescence microscopy techniques, which now offer multiple options and new opportunities for the characterization of nanostructured objects as DDSs, and of their behaviors (in terms of static localization and dynamic motion) in biological environments, on a nanometric length scale.
In this review, we summarize the major recent advancements of fluorescence microscopy-related techniques, in view of their impact in the field of drug delivery. Specifically, inSection 2, we consider advanced imaging techniques, with particular focus on super-resolution techniques (Section 2.1) and techniques designed to investigate thin layers/surfaces and thick samples (Section 2.2); in Section 3, we revise the main fluorescence microscopy-related techniques to investigate the dynamics of DDSs, with particular focus on the impact of the recent advent of super-resolution imaging on these techniques in the field of drug delivery. For the different experimental techniques, the opportunities offered by these novel tools, in relation both to DDS characterization and to the description of DDS behavior in biological environments, are summarized, highlighting the current opportunities and potentialities of static and dynamic advanced fluorescence microscopy-related techniques within drug delivery research.
2. Advanced Imaging of Drug Delivery Systems
The main application of advanced fluorescence microscopy-based techniques to the investigation of DDSs is the determination of the localization of DDSs in vitro and in vivo, which is a key issue, for instance, to: (i) understand the behavior of DDSs with biological interfaces/barriers and in biological media; (ii) evaluate the degree of cell uptake and understand cell uptake routes in vitro; and (iii) determine the extent of DDS accumulation in selected tissues. In these respects, the main advantage represented by super-resolution techniques is the possibility to accurately determine the localization of DDSs in complex biological samples. To mention a few: some super-resolution techniques necessitate the use of powerful laser sources (in particular, STED), which on one hand requires a tailored design of optimized photostable fluorescent probes, while on the other hand, might determine the photodamage and phototoxicity of biological samples in cases of long exposure [15][18]; biological samples are highly complex, therefore it is necessary to establish the efficacy/quantum yield and/or the possible inactivation of the dye in the environment of interest [16][17][18][19,20,21]; the thickness of the biological sample of interest challenged by the DDSs strongly varies from the five nanometers of a synthetic cell membrane model layered on a converglass [19][20][21][22,23,24], to the tens of micrometers of a eukaryotic cell, to the millimeters of a tissue specimen, thus requiring different tailored solutions [22][25]; in vivo imaging is extremely challenging and requires a specific setup and dedicated protocols (see, for instance, [23][26]), including the use of (relatively) biocompatible fluorescent dyes, which have to be carefully chosen when designing the experiment (in this respect, in live-cell STED microscopy, there is a common use of genetically encoded markers, such as fluorescent proteins [24][25][27,28]). In Section 2.1, the main techniques for super-resolution imaging (namely, stimulated emission depletion microscopy (STED),
In the latest years, the advent of super-resolution microscopy (SRM) has represented a game-changer in the characterization of the behavior of DDSs in biological systems—and more in general, in biomedical research—holding the promise to combine the inherent advantages of confocal microscopy (i.e., the possibility of directly visualizing biological samples without the need for complex data analysis, the non-invasive and biocompatible nature of the technique, and the possibility of highlighting specific areas of even highly complex biological samples through an appropriate selection of the fluorescent tags) with an extremely high resolution, thus approaching the limits of electron microscopy. A thorough description of the different SRM techniques is beyond the scope of this review (the reader is advised to refer to specific reviews, such as [25][26][27][28,29,30]); however, it is useful to briefly summarize the basic principles and differences of the two main experimental approaches adopted—reversible saturable optical fluorescence transitions (RESOLFT) microscopy on one side (for which the main representative technique is STED) and single-molecule localization microscopy (SMLM) on the other side (for which the main representative techniques are STORM and PALM)—in order to discuss the specific limitations and opportunities provided by diverse SRM techniques in the field of drug delivery (see Figure 1B) [28][31].
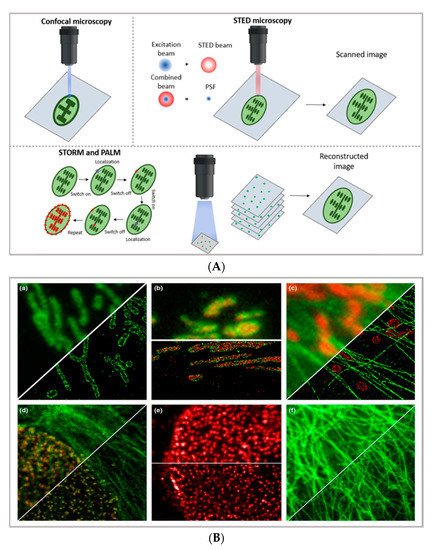
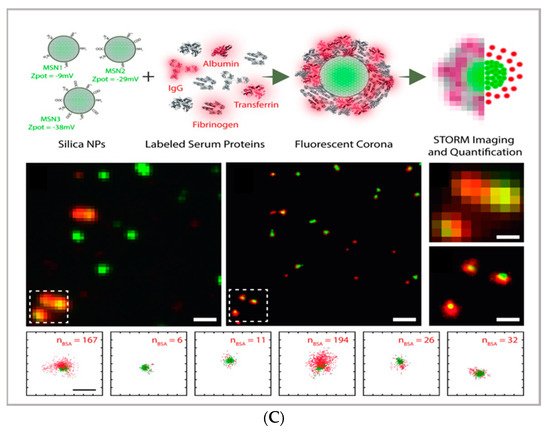
Figure 1. (A) comparison between the image acquisition/reconstruction modes of different techniques. Laser scanning confocal microscopy (LSCM) is based on a diffraction-limited point acquisition, which is scanned across the specimen through the scan-head to reconstruct the full image. Stimulated emission depletion microscopy (STED) acquisition relies on the combination of two diffraction-limited laser beams (an excitation beam and a donut-shaped depletion beam), whose combination produces a virtually unlimited decrease in the size of the point spread function; the image acquisition/reconstruction is obtained, similarly to LSCM, through a non-diffraction limited point acquisition, which is scanned across the specimen through the scan-head, to reconstruct the full image. STORM/PALM techniques rely on the employment of low concentrations of photo-switchable probes excited at a low intensity; the stochastic activation of single fluorophores allows for the determination of their precise localization, whereas repeated on-off cycles allow for the random activation of all the fluorophores in the specimen, and the combination of the cycles allows for the reconstruction of the high-resolution image [29][32]. (B) examples of high-resolution STORM and STED images compared to lower resolution LSCM images acquired for the same sample: (a–c) STORM images of BS-C-1 cells ((a,b) mitochondrial proteins Tom20 (green) and ATP Synthase (red), and (c) mitochondria protein Tom20 (red) and microtubules (green)); (d–f) multi-color STED images of PtK2 cells. All images are 10 × 10 µm in size (adapted with permission from [28][31] copyright (2015) WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany). (C) examples of super-resolution techniques applied to drug delivery research (adapted with permission from [30][33]. Scale bar 200 nm; copyright (2017) WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany).
The basic principle of STED (briefly schematized inFigure 1A) is to combine two diffraction-limited beams (an excitation beam and a donut-shaped depletion beam) to obtain a virtually unlimited decrease in the size of the point spread function (PSF). Ideally, the STED approach enables the combination of the high resolution typical of electron microscopy with the typical simpler and non-invasive sample preparation of fluorescence imaging. Moreover, the technique allows for in vivo imaging, as well as the study of subcellular interaction mechanisms and the simultaneous detection of multiple systems exploiting distinct targeting probes. STED image acquisition is relatively fast (currently, the acquisition rate of an image is limited by the scanner, rather than by the STED process); however, it requires high source power, which might be carefully considered in the application of the technique to living organisms and/or long live acquisitions of living cells.
In particular, as briefly schematized in Figure 1A,C, stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM) rely on the employment of photo-switchable/photo-activatable probes, which are dispersed within the sample in a rather diluted amount. With STORM/PALM techniques, higher spatial resolution (with lower illumination intensity than STED) is obtained, with relatively cost-effective implementations of conventional wide-field microscopes; however, a longer acquisition time and advanced image processing are required, as well as a careful choice of fluorescent probes. Conversely, fast image acquisition, without requiring complex data processing, can be achieved with STED. As a general rule of thumb in drug delivery research, SMLM techniques—with lower acquisition intensity and a slower acquisition time—can be profitably employed in the detection of relatively slow processes (such as DDS uptake and localization in living cells) with slightly higher resolution and a lower risk for phototoxicity than STED, while STED is the technique of choice for faster dynamics (as discussed inSection 3).
However, they can also represent valuable tools in characterizing complex, nanostructured DDSs; complementing the structural, colloidal, and interfacial information obtained through common characterization techniques applied to nanomaterials—such as scattering (i.e., dynamic light scattering, small-angle X-ray, and neutron scattering), surface techniques (i.e., quartz crystal microbalance with dissipation monitoring and atomic force microscopy), and electron microscopy. A similar multicolor approach has also been applied to investigate the localization and exchanges of monomers within dynamic supramolecular polymer systems, showing a possibility to exploit super-resolution microscopy to investigate the inner dynamics of multicomponent soft matter systems [31][36]. A significant opportunity in the design/synthesis of nanocarriers for drug delivery is the potential ability to functionalize the surface with targeting moieties quantitatively estimated the amount of targeting moieties (specifically, plasminogen activator inhibitor-2 (PAI-2) and trastuzumab (TZ, Herceptin®) targeting cancer cell surface biomarkers) on functionalized liposomes [32][39], showing how this experimental/analytical approach can be applied for the characterization of a multifunctional drug delivery system.
This issue also applies to fluorescence microscopy; however, in super-resolution techniques, the photophysical properties of the dyes, such as photostability, brightness, and the ability to control the ON/OFF switch of excited states, are determinant factors in the formation of super-resolved images, themselves. Therefore, in the latest years, researchers have focused on synthesizing/developing suitable fluorescent probes, especially for the appropriate labeling of complex cellular environments [15][33][34][35][36][37][38][18,40,41,42,43,44,45]. Recently, the development of nanoparticles (of an inorganic, organic, or biogenic nature) for super-resolution imaging has gained increased interest, due to the possibility of combining the new opportunities offered by super-resolution, in terms of diagnostics (with specific characteristics of nanoparticles), as therapeutic agents, or as biosensing probes [39][40][46,47]. [41][3] report on the synthesis of dye-labeled transferrin protein-based NPs with elevated photostability for super-resolution imaging in live-cell nanoscopy, thus combining the properties of transferrin-based nanoparticles (as biocompatible carriers with cancer cells’ targeting properties) with the photostability of the Atto647N dye.
3. Dynamics of Drug Delivery System
As reviewed in the previous section, thanks to the advancements of fluorescence microscopy, it is now possible to determine the localization of drug delivery with nanometric resolution. Another opportunity offered by fluorescence microscopy and laser scanning confocal microscopy-related techniques is to determine not only the localization of nanosystems or drug delivery systems but also their dynamics. In particular, these techniques can be used to: (i) characterize the cellular uptake of DDSs (which is the internalization pathway; if the DDS is internalized by cells as a whole or in a disaggregated form); (ii) characterize the binding of the DDS to relevant biomolecules; (iii) characterize the motion of the DDS in bio-relevant fluids; and (iv) characterize the interaction of the DDS with biomembranes. In the following paragraph, these techniques will be reviewed, and the opportunities provided by each technique for the field of drug delivery will be highlighted.
Specifically, PT relies on the analysis of entire fluorescence microscopy images to rebuild the trajectories of diffusing moieties. It can be applied to relatively slow processes, and it requires complex data analysis; however, it allows for the determination of very complex dynamic processes (such as those occurring in cellular uptake). It is a relatively simple technique that does not require complex experimental setup or data analysis; however, complex diffusive processes are generally difficult to disentangle, and relatively slow processes are generally considered. FCS requires a specific setup; however, it can probe very fast dynamics (such as the diffusion of a small molecule in water), can be applied to very diverse issues in drug delivery research (in the following, different examples will be presented), and requires less complex data analysis than PT—though it is less suited for disentangling highly complex diffusion modes.
In particular, by evaluating the displacement between the particle positions, it is possible to calculate the mean squared displacement (MSD) of particles linked to their diffusion coefficient. The dependence of the MSD on time provides information about the diffusion modes of the particles. For instance, Chen et al. performed real-time imaging and particle tracking with confocal microscopy to study the endocytosis and intracellular trafficking of fluorescent polymer dots and carboxylate polystyrene nanoparticles, showing the kinetics of the process at the single-particle level [42][43][44][74,75,76]. Simultaneous and real-time monitoring of the motion and intracellular dynamics of mesoporous silica nanoparticles, labeled with pH-sensitive dyes, were reported by Mou et al., who performed single particle tracking in targeting lysosomes [45][77], while Tan et al. report on the successful tracking of the endocytic transport of aptamer-drug conjugates (ApDCs) in human cancer cell lines [46][78].
A series of images is then acquired at normal illumination intensity to monitor the rate and extent of fluorescence intensity recovery inside the ROI. Specifically, from the rate of fluorescence intensity recovery (which is due to the replacement of the photobleached fluorescent dyes with active fluorescent species diffusing from the neighboring areas), the diffusion coefficient of the fluorescent species can be inferred; from the percentage of recovery compared to the theoretical fluorescence recovery, the mobile fraction of the probe is discriminated from the immobile fraction (see Figure 3b [47][69]) In drug delivery research, FRAP has been profitably applied to characterize the retention properties of hydrogels designed from drug delivery applications [48][81], as well as to determine the interaction of nanocarriers with relevant biological barriers [49][82], such as their effect on membrane fluidity [50][83]. Being a well-established technique that was developed decades ago, it is recently gaining renewed interest thanks to the possibility of coupling it with STED.
Briefly, with a laser scanning confocal microscope, the laser beam is focused on a single spot of a sample containing the fluorescent species of interest; if the fluorescently labeled species are sufficiently diluted (typically, in the nM concentration range), then their fluorescence intensity significantly fluctuates with time, due to the diffusion of the species inside and outside of the excitation volume. Additionally, by exploiting the sensitivity of the technique for the diffusion coefficient of the fluorescent species of interest (and, therefore, on its size), it has been exploited to evaluate the form (i.e., assembled or disassembled) of the DDSs inside cells [51][52][102,103]. FCS is also a powerful tool for investigating dynamic processes involving cell membranes and biomimetic membranes [53][54][105,106]. Recently, STED has been coupled with FCS, allowing for the monitoring of the dynamics of fluorescent species in spots of reduced size (i.e., below 200 nm) [55][56][107,108]; this has been applied to the monitoring of lipid dynamics [57][58][109,110].
