Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Sahar El Aidy.
A disturbed interaction between the gut microbiota and the mucosal immune system plays a pivotal role in the development of inflammatory bowel disease (IBD).
- microbial metabolites
- gut
- inflammation
1. Introduction
Inflammatory bowel disease (IBD) is an idiopathic disease affecting the gastrointestinal (GI) tract and can be divided into two main subcategories: Crohn’s disease (CD) and ulcerative colitis (UC). Both CD and UC lead to poor quality of life and psychological distress for patients, and produce significant pressure on healthcare systems by their relatively high morbidity. Genetic and environmental factors are known to increase the risk of IBD and may predispose certain individuals or populations to developing the disease. Prevalence of IBD has always been relatively high in Europe and North America, but is now also on the rise in industrializing countries in Asia, Africa, and South America [1].
Despite the lack of full understanding of the pathophysiology of IBD, the majority of available reports suggest a dysregulation between the intestinal microbiota and the host immune system (i.e., loss of immune tolerance) to be one of the underlying causes. The innate immune system in the intestinal mucosa responds to the microbiota and/or antigens by promoting inflammation, which recruits the adaptive immune system and leads to a more severe and long-lasting inflammatory state, as well as deterioration of the intestinal barrier integrity. The latter leads to translocation of microbiota and/or antigens into the mucosa, further exacerbating the mucosal inflammatory response, thereby creating a vicious circle [2,3][2][3].
Currently used pharmacological interventions are aimed at combatting the characteristic flareups of intestinal inflammation. The most effective drugs are corticosteroids and tumor necrosis factor (TNF) inhibitors. However, the former cannot be used for extended periods of time due to serious side effects (e.g., Cushing’s syndrome), and the latter has a significant amount of primary and secondary non-responders, along with serious side effects [4,5,6][4][5][6].
Fecal microbiota transplant (FMT) is another, experimental, form of IBD treatment. A recent meta-analysis found that 54% of IBD patients showed a clinical response to FMT, and 37% demonstrated clinical remission, while 29% suffered from adverse events [7]. Generally, the adverse events following FMT are mild and subside within 24 h, but more serious events, such as IBD flareups, infections, colectomy, pancreatitis, and death are also reported, although less frequently [8].
Despite the promising remission rates of this IBD treatment, which is still in its infancy, the main motive against FMT is that the treatment is considered to be a black box. The outcome and safety of the treatment is influenced by a myriad of factors (e.g., host genotype, specific type of microbiota imbalance, type and stage of IBD, route of administration, and factors related to the FMT donor), which remain obscure [8,9][8][9].
Considering the pivotal role of the gut microbiota in IBD, and that, ultimately, a major part of the communication between the gut microbiota and the host is based on chemical signaling, this review aims to examine gut microbial metabolites known to have anti-IBD effects. In order to positively implicate the role of microbial metabolism, only compounds proven to be produced by the gut microbiota have been taken into consideration. Furthermore, the metabolites discussed in this review originate from parental compounds found in common dietary sources (e.g., vegetables, fruits, and herbs), and have either been shown to improve colitis symptoms in vivo, affect signaling pathways involved in the pathophysiology of IBD in vitro, or both. The relevant data are summarized in Table 1.
Table 1. Overview of metabolites, bacterial species currently known to produce these metabolites, and experimental models used to assess anti-IBD effects.
| Microbial Metabolite | Parental Compound | Phylum | Species | Experimental Model | Ref. |
|---|
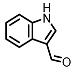 | Indole-3-aldehyde (I3Al) |
Tryptophan | Firmicutes | Lactobacillus reuteri | Lactobacillus murinus | in vitro, in vivo |
[10,11,12] | [10][11][12] | ||||||||||||||||||||||||||
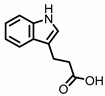 | Indole-3-propionic acid (I3Pr) |
Tryptophan | Firmicutes | Peptostreptococcus russellii | Peptostreptococcus anaerobius | Peptostreptococcus asaccharolyticus | Clostridium sporogenes | Clostridium botulinum | Clostridium caloritolerans | Clostridium paraputrificum | Clostridium cadaveris | in vitro, in vivo |
[13,14,15,16,17,18] | [13][14][15][16][17][18] | ||||||||||||||||||||
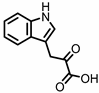 | Indole-3-pyruvic acid (I3Py) |
Tryptophan | Firmicutes | Clostridium sporogenes | in vitro, in vivo |
[15,19] | [15][19] | |||||||||||||||||||||||||||
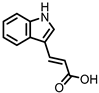 | Indole-3-acrylic acid (I3Acr) |
Tryptophan | Firmicutes | Peptostreptococcus russellii | Peptostreptococcus anaerobius | Clostridium sporogenes | in vitro | [15,20] | [15][20] | |||||||||||||||||||||||||
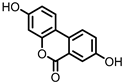 | Urolithin A (UrA) |
Ellagic acid | Actinobacteria | Bifidobacterium pseudocatenulatum | in vitro, in vivo |
[21,] | [21] | 22, | [22] | 23, | [ | 24,25, | 23][24][25] | 26 | [26] | |||||||||||||||||||
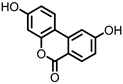 | Isouroithin A (iUrA) |
Ellagic acid | Actinobacteria | Ellagibacter isourolithinifaciens | in vitro | [25,27,28] | [25][27][28] | |||||||||||||||||||||||||||
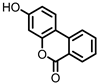 | Urolithin B (UrB) |
Ellagic acid | Actinobacteria | Bifidobacterium pseudocatenulatum | in vitro | [21,24,25] | [21][24][25] | |||||||||||||||||||||||||||
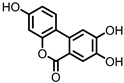 | Urolithin C (UrC) |
Ellagic acid | Actinobacteria | Gordonibacter urolithinfaciens | Gordonibacter pamelaeae | in vitro | [24,29,30] | [24][29][30] | ||||||||||||||||||||||||||
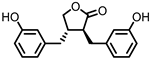 | Enterolactone (EL) | 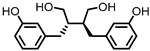 | Enterodiol (ED) |
Lignans | Firmicutes | Lactobacillus gasseri | Lactobacillus salivarius | Clostridium scindens | Lactonifactor longoviformis | Peptostreptococcus productus | in vitro | [31,32,33,34,35,36,37,38,39,40] | [31][32][33][34][35][36][37][38][39][40] | |||||||||||||||||||||
| Actinobacteria | Bifidobacterium bifidum | Bifidobacterium catenulatum | Bifidobacterium pseudolongum | Bifidobacterium adolescentis | Eggerthella lenta | |||||||||||||||||||||||||||||
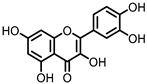 | Quercetin |
Quercitrin | Fusobacteria | Fusobacterium K-60 | in vitro, in vivo |
[41,42,43,44] | [41][42][43][44] | |||||||||||||||||||||||||||
| Rutin | Firmicutes | Enterococcus avium | Lactobacillus acidophilus | Lactobacillus plantarum | Lachnoclostridium | spp. | Eisenbergiella | spp. | Blautia | sp. | in vitro, in vivo |
[45,46,47,48,49,50,51,52,53] | [45][46][47][48][49][50][51][52][53] | |||||||||||||||||||||
| Actinobacteria | Bifidobacterium dentium | |||||||||||||||||||||||||||||||||
| Bacteroidetes | Bacteroides uniformis | Bacteroides ovatus | Parabacteroides distasonis | |||||||||||||||||||||||||||||||
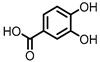 | Protocatechuic acid (PCA)/3,4-dihydroxybenzoic acid | 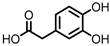 | 3,4-dihydroxyphenylacetic acid (DHPA) | 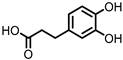 | 3,4-dihydroxyphenylpropionic acid (DHPP) |
Flavonols Flavan-3-ols Flavones Anthocyanins |
Firmicutes | Eubacterium oxidoreducens | Eubacterium ramulus | Enterococcus casseliflavus | Flavonifractor plautii | Catenibacillus scindens | Butyrivibrio | spp. | in vitro, in vivo |
[54,55,56,57,58,59,60,61,62, | 55][56][57] | 63, | [ | 64,65, | 58][ | 66, | 59][[62][63][64][65][66] | 67, | 60][ | 68, | [67] | 69] | [54][61][68][69] | |||||
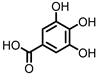 | Gallic acid (GA)/3,4,5-trihydroxybenzoic acid |
Anthocyanins | Firmicutes | Lactobacillus plantarum | Lactobacillus casei | in vitro, in vivo |
[70,71,72,] | [70] | 73, | [71] | 74, | [ | 75,76, | 72][73][74][75][76] | 77 | [77] | ||||||||||||||||||
| Actinobacteria | Bifidobacterium lactis | |||||||||||||||||||||||||||||||||
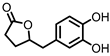 | 3,4-dihydroxyphenyl-γ-valeric lactone (DHPVL) |
Flavan-3-ols Proanthocyanins |
Firmicutes | Lactobacillus plantarum | Clostridium coccoides | Flavonifractor plautii | in vitro | [54,55,58,59, | ][59 | 60, | ][60 | 63, | ][63 | 78, | ][78 | 79,80] | [54][55][58][79][80] | |||||||||||||||||
| Actinobacteria | Eggerthella lenta | Eggerthella | sp. | |||||||||||||||||||||||||||||||
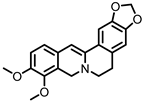 | Dihydroberberine |
Berberine | Firmicutes | Enterococcus faecium | Enterococcus faecalis | Staphylococcus aureus | Staphylococcus epidermis | in vitro | a | , in vivo | [81,82,83,84,85,86,87] | [81][82][83][84][85][86][87] | ||||||||||||||||||||||
| Proteobacteria | Escherichia coli | Enterobacter cloacae | Klebsiella pneumoniae | |||||||||||||||||||||||||||||||
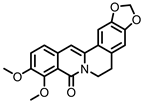 | Oxyberberine |
Berberine | Firmicutes | Lactobacillus acidophilus | Streptococcus aureus | in vivo | [88] | |||||||||||||||||||||||||||
| Actinobacteria | Bifidobacterium longum | |||||||||||||||||||||||||||||||||
| Proteobacteria | Escherichia coli | Pseudomonas aeruginosa | ||||||||||||||||||||||||||||||||
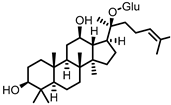 | Compound K (CK) |
Ginsenoside Rb1 | Firmicutes | Eubacterium | in vitro, in vivo |
[89,90,91,92,93,94] | [89][90][91][92][93][94] | |||||||||||||||||||||||||||
| Actinobacteria | Bifidobacterium | |||||||||||||||||||||||||||||||||
| Bacteroidetes | Bacteroides | |||||||||||||||||||||||||||||||||
| Fusobacteria | Fusobacterium |
Due to intrinsic differences in the interindividual dietary and microbiota compositions, especially the disturbed microbiota of IBD patients, such metabolites may not be produced universally. Identifying these metabolites can help to overcome such intrinsic differences, and, ideally, helps making gut health less dependent on changes in the microbiota composition.
2. Indoles
Indole derivatives (Figure 1) are mainly produced by Lactobacilli, Clostridia, Peptostreptococci, Bifidobacteria, and Bacteroides (Table 1), as metabolites of the amino acid tryptophan (Trp) [95]. Gut microbial Trp metabolites are often found to be agonists of the aryl hydrocarbon receptor (AHR), of which lower levels are observed in IBD patients, compared to healthy subjects [96]. IBD symptoms and pro-inflammatory cytokine levels were found to be greater in AHR knockouts in murine models of dextran sodium sulfate (DSS)-induced colitis [97]. Other AHR ligands are known to reduce colitis symptoms [96,98][96][98].
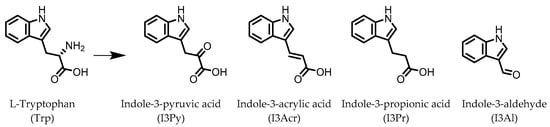
Figure 1.
Structures of L-tryptophan and several indole metabolites produced by the gut microbiota.
AHR activation by the gut microbial Trp metabolite indole-3-aldehyde (I3Al) was shown to stimulate mucosal lymphocytes to secrete interleukin 22 (IL-22), an anti-inflammatory cytokine known to play an important role in protecting mice from developing IBD [99]. Increased IL-22 secretion causes signal transducer and activator of transcription 3 (STAT3) phosphorylation, which ultimately leads to faster proliferation of intestinal epithelial cells (IECs), contributing to the recovery of damaged intestinal mucosa following DSS-induced colitis [10].
Indole-3-propionic acid (I3Pr) also activates the AHR receptor, which induced IL-10 receptor expression in cultured IECs. Oral administration of I3Pr was shown to improve DSS-induced murine colitis symptoms, which was attributed to increased signaling of the anti-inflammatory cytokine IL-10, due to higher expression of IL-10 receptors [13].
Additionally, I3Pr was found to act as a ligand for the pregnane X receptor (PXR) in vivo, and led to lower TNF-α levels together with higher levels of mRNA coding for tight junction proteins, thus contributing to intestinal integrity. With the help of knockout experiments, it was determined that activation of PXR modulates Toll-like receptor 4 (TLR4) signaling, which is known to activate nuclear factor κB (NF-κB), a pro-inflammatory transcription factor. Accordingly, oral administration of I3Pr could activate PXR in the colon, which prevents lipopolysaccharide (LPS)-induced inflammation via modulation of TLR4, thereby preserving the intestinal integrity [14].
Administration of indole-3-pyruvic acid (I3Py) to mice with CD4+ T cell-induced colitis led to an increase in the amount of IL-10-producing T cells, while the number of Th1 cells in the mucosa was decreased, resulting in a reduction in colitis symptoms [19].
In a co-culture of murine-derived colonic spheroids and murine bone marrow-derived macrophages (BMDMs), indole-3-acrylic acid (I3Acr) promoted IL-10 secretion while suppressing TNF-α production upon stimulation with LPS, via activation of AHR. This stimulated the expression of the mucin protein coding gene, Muc2, which may help to protect the intestinal epithelium. When human peripheral blood mononuclear cells (PBMCs) were treated with I3Acr, a reduction in IL-1β and IL-6 was observed, upon LPS stimulation. Moreover, not only was AHR activation reproduced in the human cell line, activation of the anti-inflammatory Nrf2–ARE pathway was observed. Using these human PBMCs in the co-culture, I3Acr treatment promoted important anti-inflammatory and anti-oxidant effects, by upregulating Nrf2- and AHR-pathway target genes and genes related to the biosynthesis glutathione (GSH), an important anti-oxidant that protects cells from oxidative stress [20].
3. Urolithins
Urolithins are gut microbial metabolites of ellagic acid, a hydrolysis product of ellagitannins (Figure 2). Both ellagic acid and ellagitannins are naturally found in various fruits, nuts, and seeds (e.g., pomegranate, raspberry, strawberry, almond, and walnut) [100]. Several members of the Actinobacteria (Table 1) have been found to metabolize ellagic acid into particular urolithins, which differ by the number and the positions of hydroxyl groups. For example, Gordonibacter urolithinfaciens and Gordonibacter pamelaeae are able to produce urolithin C (UrC), but are not capable of further dehydroxylation [29,30][29][30]. Urolithin A (UrA) and urolithin B (UrB) are produced by Bifidobacterium pseudocatenulatum, whereas isourolithin A (iUrA) is produced by Ellagibacter isourolithinifaciens [21,27,28][21][27][28].
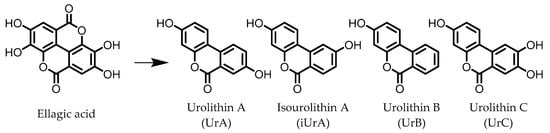
Figure 2.
Structures of ellagic acid and several urolithins produced by the gut microbiota.
A comparison between the effects of pomegranate extract (PE) and UrA on DSS-induced colitis in rats showed that both were able to decrease levels of the pro-inflammatory mediators nitric oxide (NO) and prostaglandin E2 (PGE2) in colonic mucosa, by downregulating the enzymes responsible for their production: inducible nitric oxide synthase (iNOS), cyclooxygenase 2 (COX-2), and prostaglandin E synthase (PTGES). However, only in the case of UrA administration was the colonic architecture protected. Additionally, UrA was able to significantly downregulate the pro-inflammatory cytokines IL-1β and IL-4, and cluster of differentiation 40 (CD40), a receptor protein involved in immune and inflammatory signaling pathways [22].
It was also observed that less UrA was produced from PE in colitic rats compared to healthy rats, suggesting that UrA production from gut microbiota, which might be absent in inflammation, plays a protective role against colitis. During colitis, UrA itself had to be administered in order to benefit from the anti-inflammatory effects. Another protective effect of UrA might be via an observed increase in the abundance of Lactobacilli, Bifidobacteria, and Clostridia taxa, which have been shown to prevent inflammation in IECs in response to pathogenic Enterobacteria [101]. Moreover, an increase in E. coli, observed after DSS treatment, was found to be lower in the rats that received UrA [22].
Several in vitro studies have been performed in an attempt to reveal a more detailed mechanism explaining the anti-inflammatory actions of UrA. The production of pro-inflammatory mediators was strongly reduced by UrA in LPS-stimulated RAW264 macrophages. UrA was found to inhibit the phosphorylation of protein kinase B (Akt) and c-Jun, effectively suppressing the pro-inflammatory PI3-K/Akt/NF-κB and JNK/AP-1 signaling pathways. This meant the downstream production of pro-inflammatory mediators (TNF-α, IL-6, and NO) was also suppressed. Notably, UrA appeared to also inhibit NADPH oxidase (NOX), which is largely responsible for production of reactive oxygen species (ROS) in activated macrophages, presenting another possible mechanism for inhibiting the activation of the pro-inflammatory transcription factors NF-κB and AP-1 [23].
iUrA, UrB, and UrC also display anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages, although the effects are inferior to UrA. The urolithins were shown to decrease the DNA-binding activity of the NF-κB p50 subunit, as well as the nuclear translocation of the p65 subunit, resulting in lower levels of TNF-α, IL-1β, IL-6, iNOS, and NO [24,25][24][25]. Additionally, UrA has been shown to promote anti-inflammatory effects in human macrophages and neutrophils, which was attributed to an observed induction of extracellular signal-regulated kinase 1 and 2 (ERK1/2) phosphorylation [25].
Besides the anti-inflammatory properties and modulation of the microbiota, UrA can also improve gut health by enhancing the intestinal barrier function. UrA was shown to activate AHR and Nrf2, which leads to the upregulation of the tight junction proteins claudin 4, occludin, and zonula occludens-1 (ZO-1). Treatment with UrA decreased gut permeability in mice with 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis, and reduced both local and systemic inflammation. When UrA was administered prior to TNBS-administration, the development of colitis was prevented. Finally, chronic and acute DSS-induced colitis were ameliorated by UrA treatment [26].
References
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778.
- Guan, Q. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019, 2019, 1–16.
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin. Proc. 2019, 94, 155–165.
- Waljee, A.K.; Wiitala, W.L.; Govani, S.; Stidham, R.; Saini, S.; Hou, J.; Feagins, L.A.; Khan, N.; Good, C.B.; Vijan, S.; et al. Corticosteroid Use and Complications in a US Inflammatory Bowel Disease Cohort. PLoS ONE 2016, 11, e0158017.
- Chudy-Onwugaje, K.O.; Christian, K.E.; Farraye, F.A.; Cross, R.K. A State-of-the-Art Review of New and Emerging Therapies for the Treatment of IBD. Inflamm. Bowel Dis. 2019, 25, 820–830.
- Hazel, K.; O’Connor, A. Emerging treatments for inflammatory bowel disease. Ther. Adv. Chronic Dis. 2020, 11.
- Caldeira, L.D.F.; Borba, H.H.; Tonin, F.S.; Wiens, A.; Fernandez-Llimos, F.; Pontarolo, R. Fecal microbiota transplantation in inflammatory bowel disease patients: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0238910.
- Basso, P.J.; Câmara, N.O.S.; Sales-Campos, H. Microbial-Based Therapies in the Treatment of Inflammatory Bowel Disease—An Overview of Human Studies. Front. Pharmacol. 2019, 9, 1571.
- Tan, P.; Li, X.; Shen, J.; Feng, Q. Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease: An Update. Front. Pharmacol. 2020, 11, 1–8.
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.R.; Yu, Q. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018, 25, 1657–1670.
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mähler, A.; Balogh, A.; Markó, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589.
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385.
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194.
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 2014, 41, 296–310.
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nat. Cell Biol. 2017, 551, 648–652.
- Elsden, S.R.; Hilton, M.G.; Waller, J.M. The end products of the metabolism of aromatic amino acids by clostridia. Arch. Microbiol. 1976, 107, 283–288.
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703.
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302.
- Aoki, R.; Aoki-Yoshida, A.; Suzuki, C.; Takayama, Y. Indole-3-Pyruvic Acid, an Aryl Hydrocarbon Receptor Activator, Suppresses Experimental Colitis in Mice. J. Immunol. 2018, 201, 3683–3693.
- Wlodarska, M.; Luo, C.; Kolde, R.; D’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6.
- Gaya, P.; Peirotén, Á.; Medina, M.; Álvarez, I.; Landete, J.M. Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins A and B from ellagic acid. J. Funct. Foods 2018, 45, 95–99.
- Larrosa, M.; González-Sarrías, A.; Yáñez-Gascón, M.J.; Selma, M.V.; Azorín-Ortuño, M.; Toti, S.; Tomás-Barberán, F.; Dolara, P.; Espín, J.C. Anti-inflammatory properties of a pomegranate extract and its metabolite urolithin-A in a colitis rat model and the effect of colon inflammation on phenolic metabolism. J. Nutr. Biochem. 2010, 21, 717–725.
- Komatsu, W.; Kishi, H.; Yagasaki, K.; Ohhira, S. Urolithin A attenuates pro-inflammatory mediator production by suppressing PI3-K/Akt/NF-κB and JNK/AP-1 signaling pathways in lipopolysaccharide-stimulated RAW264 macrophages: Possible involvement of NADPH oxidase-derived reactive oxygen species. Eur. J. Pharmacol. 2018, 833, 411–424.
- Piwowarski, J.P.; Kiss, A.K.; Granica, S.; Moeslinger, T. Urolithins, gut microbiota-derived metabolites of ellagitannins, inhibit LPS-induced inflammation in RAW 264.7 murine macrophages. Mol. Nutr. Food Res. 2015, 59, 2168–2177.
- Bobowska, A.; Granica, S.; Filipek, A.; Melzig, M.F.; Moeslinger, T.; Zentek, J.; Kruk, A.; Piwowarski, J.P. Comparative studies of urolithins and their phase II metabolites on macrophage and neutrophil functions. Eur. J. Nutr. 2020, 1–16.
- Singh, R.; Chandrashekharappa, S.; Bodduluri, S.R.; Baby, B.V.; Hegde, B.; Kotla, N.G.; Hiwale, A.A.; Saiyed, T.; Patel, P.; Vijay-Kumar, M.; et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 2019, 10, 1–18.
- Selma, M.V.; Beltrán, D.; Luna, M.C.; Vaquero, M.R.; García-Villalba, R.; Mira, A.; Espín, J.C.; Tomás-Barberán, F.A. Isolation of Human Intestinal Bacteria Capable of Producing the Bioactive Metabolite Isourolithin A from Ellagic Acid. Front. Microbiol. 2017, 8, 1521.
- Beltrán, D.; Romo-Vaquero, M.; Espín, J.C.; Tomás-Barberán, F.A.; Selma, M.V. Ellagibacter isourolithinifaciens gen. nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut. Int. J. Syst. Evol. Microbiol. 2018, 68, 1707–1712.
- Selma, M.V.; Beltrán, D.; García-Villalba, R.; Espín, J.C.; Tomás-Barberán, F.A. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014, 5, 1779–1784.
- Selma, M.V.; Tomás-Barberán, F.A.; Beltrán, D.; García-Villalba, R.; Espín, J.C. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int. J. Syst. Evol. Microbiol. 2014, 64, 2346–2352.
- Jin, J.-S.; Zhao, Y.-F.; Nakamura, N.; Akao, T.; Kakiuchi, N.; Min, B.-S.; Hattori, M. Enantioselective Dehydroxylation of Enterodiol and Enterolactone Precursors by Human Intestinal Bacteria. Biol. Pharm. Bull. 2007, 30, 2113–2119.
- Peirotén, Á.; Gaya, P.; Álvarez, I.; Bravo, D.; Landete, J.M. Influence of different lignan compounds on enterolignan production by Bifidobacterium and Lactobacillus strains. Int. J. Food Microbiol. 2019, 289, 17–23.
- Gaya, P.; Peirotén, Á.; Medina, M.; Landete, J.M. Bifidobacterium adolescentis INIA P784: The first probiotic bacterium capable of producing enterodiol from lignan extracts. J. Funct. Foods 2017, 29, 269–274.
- Bravo, D.; Peirotén, Á.; Álvarez, I.; Landete, J.M. Phytoestrogen metabolism by lactic acid bacteria: Enterolignan production by Lactobacillus salivarius and Lactobacillus gasseri strains. J. Funct. Foods 2017, 37, 373–378.
- Clavel, T.; Lippman, R.; Gavini, F.; Doré, J.; Blaut, M. Clostridium saccharogumia sp. nov. and Lactonifactor longoviformis gen. nov., sp. nov., two novel human faecal bacteria involved in the conversion of the dietary phytoestrogen secoisolariciresinol diglucoside. Syst. Appl. Microbiol. 2007, 30, 16–26.
- Clavel, T.; Henderson, G.; Engst, W.; Dorã, J.; Blaut, M.; Doré, J. Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol. Ecol. 2006, 55, 471–478.
- Clavel, T.; Borrmann, D.; Braune, A.; Doré, J.; Blaut, M. Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe 2006, 12, 140–147.
- Wang, L.-Q.; Meselhy, M.R.; Li, Y.; Qin, G.-W.; Hattori, M. Human Intestinal Bacteria Capable of Transforming Secoisolariciresinol Diglucoside to Mammalian Lignans, Enterodiol and Enterolactone. Chem. Pharm. Bull. 2000, 48, 1606–1610.
- Corsini, E.; Dell’Agli, M.; Facchi, A.; De Fabiani, E.; Lucchi, L.; Boraso, M.S.; Marinovich, M.; Galli, C.L. Enterodiol and Enterolactone Modulate the Immune Response by Acting on Nuclear Factor-κB (NF-κB) Signaling. J. Agric. Food Chem. 2010, 58, 6678–6684.
- Almousa, A.A.; Meurens, F.; Krol, E.S.; Alcorn, J. Linoorbitides and enterolactone mitigate inflammation-induced oxidative stress and loss of intestinal epithelial barrier integrity. Int. Immunopharmacol. 2018, 64, 42–51.
- Camuesco, D.; Comalada, M.; Rodriguez-Cabezas, M.E.; Nieto, A.; Lorente, M.D.; Concha, A.; Zarzuelo, A.; Gálvez, J. The intestinal anti-inflammatory effect of quercitrin is associated with an inhibition in iNOS expression. Br. J. Pharmacol. 2004, 143, 908–918.
- Comalada, M.; Camuesco, D.; Sierra, S.; Ballester, I.; Xaus, J.; Gálvez, J.; Zarzuelo, A. In vivoquercitrin anti-inflammatory effect involves release of quercetin, which inhibits inflammation through down-regulation of the NF-κB pathway. Eur. J. Immunol. 2005, 35, 584–592.
- Kim, D.-H.; Kim, S.-Y.; Park, S.-Y.; Han, M.J. Metabolism of Quercitrin by Human Intestinal Bacteria and Its Relation to Some Biological Activities. Biol. Pharm. Bull. 1999, 22, 749–751.
- Park, S.Y.; Kim, J.H.; Kim, D.H. Purification and characterization of quercitrin-hydrolyzing α-L-rhamnosidase from Fusobacterium K-60, a human intestinal bacterium. J. Microbiol. Biotechnol. 2005, 15, 519–524.
- Mascaraque, C.; Aranda, C.; Ocón, B.; Monte, M.J.; Suárez, M.D.; Zarzuelo, A.; Marín, J.J.G.; Martínez-Augustin, O.; de Medina, F.S. Rutin has intestinal antiinflammatory effects in the CD4+ CD62L+ T cell transfer model of colitis. Pharmacol. Res. 2014, 90, 48–57.
- Kwon, K.H.; Murakami, A.; Tanaka, T.; Ohigashi, H. Dietary rutin, but not its aglycone quercetin, ameliorates dextran sulfate sodium-induced experimental colitis in mice: Attenuation of pro-inflammatory gene expression. Biochem. Pharmacol. 2005, 69, 395–406.
- Cruz, T.; Gálvez, J.; Ocete, M.; Crespo, M.; de Medina, F.-H.S.; Zarzuelo, A. Oral administration of rutoside can ameliorate inflammatory bowel disease in rats. Life Sci. 1998, 62, 687–695.
- Bokkenheuser, V.D.; Shackleton, C.H.; Winter, J. Hydrolysis of dietary flavonoid glycosides by strains of intestinal Bacteroides from humans. Biochem. J. 1987, 248, 953–956.
- Seo-Hyeon, B.; Yang-Jin, H.; Juwon, S.; Sung-Woon, H.; Dong-Hyun, K. Metabolism of Rutin and Poncirin by Human Intestinal Microbiota and Cloning of Their Metabolizing. J. Microbiol. Biotechnol. 2015, 25, 18–25.
- Shin, N.R.; Moon, J.S.; Shin, S.-Y.; Li, L.; Lee, Y.B.; Kim, T.-J.; Han, N.S. Isolation and characterization of human intestinal Enterococcus avium EFEL009 converting rutin to quercetin. Lett. Appl. Microbiol. 2016, 62, 68–74.
- Beekwilder, J.; Marcozzi, D.; Vecchi, S.; De Vos, R.; Janssen, P.; Francke, C.; Vlieg, J.V.H.; Hall, R.D. Characterization of Rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl. Environ. Microbiol. 2009, 75, 3447–3454.
- Riva, A.; Kolimár, D.; Spittler, A.; Wisgrill, L.; Herbold, C.W.; Abrankó, L.; Berry, D. Conversion of Rutin, a Prevalent Dietary Flavonol, by the Human Gut Microbiota. Front. Microbiol. 2020, 11, 1–11.
- Kim, M.; Kim, N.; Han, J. Metabolism of Kaempferia parviflora Polymethoxyflavones by Human Intestinal Bacterium Bautia sp. MRG-PMF1. J. Agric. Food Chem. 2014, 62, 12377–12383.
- Corrêa, T.A.F.; Rogero, M.M.; Hassimotto, N.M.A.; Lajolo, F.M. The Two-Way Polyphenols-Microbiota Interactions and Their Effects on Obesity and Related Metabolic Diseases. Front. Nutr. 2019, 6, 188.
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234.
- Jang, S.-E.; Choi, J.-R.; Han, M.J.; Kim, N.-H. The Preventive and Curative Effect of Cyanidin-3β-D-Glycoside and Its Metabolite Protocatechuic Acid Against TNBS-induced Colitis in Mice. Nat. Prod. Sci. 2016, 22, 282–286.
- Min, S.-W.; Ryu, S.-N.; Kim, D.-H. Anti-inflammatory effects of black rice, cyanidin-3-O-β-d-glycoside, and its metabolites, cyanidin and protocatechuic acid. Int. Immunopharmacol. 2010, 10, 959–966.
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The Reciprocal Interactions between Polyphenols and Gut Microbiota and Effects on Bioaccessibility. Nutrients 2016, 8, 78.
- Moco, S.; Martin, F.-P.J.; Rezzi, S. Metabolomics View on Gut Microbiome Modulation by Polyphenol-rich Foods. J. Proteome Res. 2012, 11, 4781–4790.
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24.
- Braune, A.; Blaut, M. Deglycosylation of puerarin and other aromatic C-glucosides by a newly isolated human intestinal bacterium. Environ. Microbiol. 2011, 13, 482–494.
- Braune, A.; Blaut, M. Catenibacillus scindens gen. nov., sp. nov., a C-deglycosylating human intestinal representative of the Lachnospiraceae. Int. J. Syst. Evol. Microbiol. 2018, 68, 3356–3361.
- Marín, L.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Bioavailability of Dietary Polyphenols and Gut Microbiota Metabolism: Antimicrobial Properties. BioMed Res. Int. 2015, 2015, 905215.
- Farombi, E.O.; Adedara, I.A.; Awoyemi, O.V.; Njoku, C.R.; Micah, G.O.; Esogwa, C.U.; Owumi, S.E.; Olopade, J.O. Dietary protocatechuic acid ameliorates dextran sulphate sodium-induced ulcerative colitis and hepatotoxicity in rats. Food Funct. 2016, 7, 913–921.
- Crespo, I.; San-Miguel, B.; Mauriz, J.L.; de Urbina, J.O.; Almar, M.; Tuñón, M.J.; González-Gallego, J. Protective Effect of Protocatechuic Acid on TNBS-Induced Colitis in Mice Is Associated with Modulation of the SphK/S1P Signaling Pathway. Nutrients 2017, 9, 288.
- Hu, R.; He, Z.; Liu, M.; Tan, J.; Zhang, H.; Hou, D.-X.; He, J.; Wu, S. Dietary protocatechuic acid ameliorates inflammation and up-regulates intestinal tight junction proteins by modulating gut microbiota in LPS-challenged piglets. J. Anim. Sci. Biotechnol. 2020, 11, 1–12.
- Larrosa, M.; Luceri, C.; Vivoli, E.; Pagliuca, C.; Lodovici, M.; Moneti, G.; Dolara, P. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol. Nutr. Food Res. 2009, 53, 1044–1054.
- Monagas, M.; Khan, N.; Andrés-Lacueva, C.; Urpí-Sardá, M.; Vázquez-Agell, M.; Lamuela-Raventós, R.M.; Estruch, R. Dihydroxylated phenolic acids derived from microbial metabolism reduce lipopolysaccharide-stimulated cytokine secretion by human peripheral blood mononuclear cells. Br. J. Nutr. 2009, 102, 201–206.
- Miene, C.; Weise, A.; Glei, M. Impact of Polyphenol Metabolites Produced by Colonic Microbiota on Expression of COX-2 and GSTT2 in Human Colon Cells (LT97). Nutr. Cancer 2011, 63, 653–662.
- Pandurangan, A.K.; Mohebali, N.; Esa, N.M.; Looi, C.Y.; Ismail, S.; Saadatdoust, Z. Gallic acid suppresses inflammation in dextran sodium sulfate-induced colitis in mice: Possible mechanisms. Int. Immunopharmacol. 2015, 28, 1034–1043.
- Pandurangan, A.K.; Mohebali, N.; Norhaizan, M.E.; Looi, C.Y. Gallic acid attenuates dextran sulfate sodium-induced experimental colitis in BALB/c mice. Drug Des. Dev. Ther. 2015, 9, 3923–3934.
- Bayramoglu, A.; Kanbak, G.; Canbek, M.; Dokumac, E. Gallic acid Reduces Experimental Colitis in Rats by Downregulation of Cathepsin and Oxidative Stress. Erciyes Med J. 2020, 42, 213–217.
- Khodayar, B.; Farzaei, M.H.; Hossein Abdolghaffari, A.; Bahramsoltani, R.; Baeeri, M.; Sabbagh Ziara-ni, F.; Mohammadi, M.; Rahimi, R.; Abdollahi, M. The Protective Effect of the Gallic Acid Against TNBS-induced Ulcerative Colitis in Rats: Role of Inflammatory Parameters. J. Iran. Med. Counc. 2018, 1, 34–42.
- Zhu, L.; Gu, P.; Shen, H. Gallic acid improved inflammation via NF-κB pathway in TNBS-induced ulcerative colitis. Int. Immunopharmacol. 2019, 67, 129–137.
- Marinov, V.P.; Tzaneva, M.A.; Zhelyazkova-Savova, M.D.; Gancheva, S.; Valcheva-Kuzmanova, S.V. Effects of gallic acid in a rat model of inflammatory bowel disease induced by trinitrobenzenesulfonic acid. Bulg. Chem. Commun. 2019, 51, 22–28.
- Ávila, M.; Hidalgo, M.; Sánchez-Moreno, C.; Pelaez, C.; Requena, T.; de Pascual-Teresa, S. Bioconversion of anthocyanin glycosides by Bifidobacteria and Lactobacillus. Food Res. Int. 2009, 42, 1453–1461.
- Hidalgo, M.; Oruna-Concha, M.J.; Kolida, S.; Walton, G.E.; Kallithraka, S.; Spencer, J.P.E.; Gibson, G.R.; De Pascual-Teresa, S. Metabolism of Anthocyanins by Human Gut Microflora and Their Influence on Gut Bacterial Growth. J. Agric. Food Chem. 2012, 60, 3882–3890.
- Uhlenhut, K.; Högger, P. Facilitated cellular uptake and suppression of inducible nitric oxide synthase by a metabolite of maritime pine bark extract (Pycnogenol). Free Radic. Biol. Med. 2012, 53, 305–313.
- Sun, Y.N.; Li, W.; Song, S.B.; Yan, X.T.; Zhao, Y.; Jo, A.R.; Kang, J.S.; Ho, K.Y. A new phenolic derivative with soluble epoxide hydrolase and nuclear factor-kappaB inhibitory activity from the aqueous extract of Acacia catechu. Nat. Prod. Res. 2016, 30, 2085–2092.
- Kim, H.S.; Chung, S.; Song, M.-Y.; Lim, C.; Shin, H.; Hur, J.; Kwon, H.; Suh, Y.-G.; Kim, E.-H.; Shin, D.; et al. Efficient and Divergent Enantioselective Syntheses of DHPVs and Anti-Inflammatory Effect on IEC-6 Cells. Molecules 2020, 25, 2215.
- Gu, L.; Li, N.; Li, Q.; Zhang, Q.; Wang, C.; Zhu, W.; Li, J. The effect of berberine in vitro on tight junctions in human Caco-2 intestinal epithelial cells. Fitoterapia 2009, 80, 241–248.
- Li, N.; Gu, L.; Qu, L.; Gong, J.; Li, Q.; Zhu, W.; Li, J. Berberine attenuates pro-inflammatory cytokine-induced tight junction disruption in an in vitro model of intestinal epithelial cells. Eur. J. Pharm. Sci. 2010, 40, 1–8.
- Gu, L.; Li, N.; Gong, J.; Li, Q.; Zhu, W.; Li, J. Berberine Ameliorates Intestinal Epithelial Tight-Junction Damage and Down-regulates Myosin Light Chain Kinase Pathways in a Mouse Model of Endotoxinemia. J. Infect. Dis. 2011, 203, 1602–1612.
- Takahara, M.; Takaki, A.; Hiraoka, S.; Adachi, T.; Shimomura, Y.; Matsushita, H.; Nguyen, T.T.T.; Koike, K.; Ikeda, A.; Takashima, S.; et al. Berberine improved experimental chronic colitis by regulating interferon-γ- and IL-17A-producing lamina propria CD4+ T cells through AMPK activation. Sci. Rep. 2019, 9, 1–13.
- Li, H.; Feng, C.; Fan, C.; Yang, Y.; Yang, X.; Lu, H.; Lu, Q.; Zhu, F.; Xiang, C.; Zhang, Z.; et al. Intervention of oncostatin M-driven mucosal inflammation by berberine exerts therapeutic property in chronic ulcerative colitis. Cell Death Dis. 2020, 11, 1–17.
- Jia, L.; Xue, K.; Liu, J.; Habotta, O.A.; Hu, L.; Moneim, A.E.A.; Caccamo, D. Anticolitic Effect of Berberine in Rat Experimental Model: Impact of PGE2/p38 MAPK Pathways. Mediat. Inflamm. 2020, 2020, 1–12.
- Feng, R.; Shou, J.-W.; Zhao, Z.-X.; He, C.-Y.; Ma, C.; Huang, M.; Fu, J.; Tan, X.-S.; Li, X.-Y.; Wen, B.-Y.; et al. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci. Rep. 2015, 5, 12155.
- Li, C.; Ai, G.; Wang, Y.; Lu, Q.; Luo, C.; Tan, L.; Lin, G.; Liu, Y.; Li, Y.; Zeng, H.; et al. Oxyberberine, a novel gut microbiota-mediated metabolite of berberine, possesses superior anti-colitis effect: Impact on intestinal epithelial barrier, gut microbiota profile and TLR4-MyD88-NF-κB pathway. Pharmacol. Res. 2020, 152, 104603.
- Kim, D.-H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 2018, 42, 255–263.
- Yang, L.; Zou, H.; Gao, Y.; Luo, J.; Xie, X.; Meng, W.; Zhou, H.; Tan, Z. Insights into gastrointestinal microbiota-generated ginsenoside metabolites and their bioactivities. Drug Metab. Rev. 2020, 52, 125–138.
- Wang, C.-Z.; Yao, H.; Zhang, C.-F.; Chen, L.; Wan, J.-Y.; Huang, W.-H.; Zeng, J.; Zhang, Q.-H.; Liu, Z.; Yuan, J.; et al. American ginseng microbial metabolites attenuate DSS-induced colitis and abdominal pain. Int. Immunopharmacol. 2018, 64, 246–251.
- Joh, E.-H.; Lee, I.-A.; Jung, I.-H.; Kim, D.-H. Ginsenoside Rb1 and its metabolite compound K inhibit IRAK-1 activation—The key step of inflammation. Biochem. Pharmacol. 2011, 82, 278–286.
- Li, J.; Zhong, W.; Wang, W.; Hu, S.; Yuan, J.; Zhang, B.; Hu, T.; Song, G. Ginsenoside Metabolite Compound K Promotes Recovery of Dextran Sulfate Sodium-Induced Colitis and Inhibits Inflammatory Responses by Suppressing NF-κB Activation. PLoS ONE 2014, 9, e87810.
- Zhang, J.; Cao, L.; Wang, H.; Cheng, X.; Wang, L.; Zhu, L.; Yan, T.; Xie, Y.; Wu, Y.; Zhao, M.; et al. Ginsenosides Regulate PXR/NF-κB Signaling and Attenuate Dextran Sulfate Sodium-Induced Colitis. Drug Metab. Dispos. 2015, 43, 1181–1189.
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 1–10.
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; Macdonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals Up-regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, 237–248.e1.
- Wang, Q.; Yang, K.; Han, B.; Sheng, B.; Yin, J.; Pu, A.; Li, L.; Sun, L.; Yuan, Q.; Kunqiu, Y.; et al. Aryl hydrocarbon receptor inhibits inflammation in DSS-induced colitis via the MK2/p-MK2/TTP pathway. Int. J. Mol. Med. 2018, 41, 868–876.
- Neavin, D.R.; Liu, D.; Ray, B.; Weinshilboum, R.M. The Role of the Aryl Hydrocarbon Receptor (AHR) in Immune and Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 3851.
- Zenewicz, L.A.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.; Stevens, S.; Flavell, R.A. Innate and Adaptive Interleukin-22 Protects Mice from Inflammatory Bowel Disease. Immunity 2008, 29, 947–957.
- Landete, J. Ellagitannins, ellagic acid and their derived metabolites: A review about source, metabolism, functions and health. Food Res. Int. 2011, 44, 1150–1160.
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292.
More
