Fe and P deficiency responses, which enable plants to improve their ability to acquire these nutrients, show many similarities.
Iron (Fe) and phosphorus (P) are two essential elements for plant growth whose nutrition is closely related. Both elements are abundant in soils but with poor availability for plants, which favor their acquisition by developing root morphological and physiological responses. Ethylene (ET) and nitric oxide (NO) have been involved in the regulation of both Fe and P related acquisition genes. The common involvement of ET and NO suggests that they must act in conjunction with other specific signals, more closely related to each deficiency, to avoid the induction of the specific responses when ET/NO increase due to other nutrient deficiencies or stresses. Besides the specificity conferred by these signals, ET itself could confer specificity to the responses to Fe- or P-deficiency by acting through different signaling pathways in each case.
- iron
- phosphorous
- nutrient deficiencies
- Strategy I
- ethylene
- nitric oxide
- S-nitrosoglutathione
- acid phosphatase
- ferric reductase
- EIN3
1. Fe and P acquisition by Dicotyledonous Plants
To acquire Fe, dicotyledonous plants use Strategy I [16][1], which is characterized by the necessity of reducing Fe3+, the most abundant form in soil, to Fe2+, prior to its absorption. This reduction is mediated by a ferric reductase located in the plasma membrane of the epidermal root cells and encoded by FRO2 in Arabidopsis [17] [2]. Once Fe3+ has been reduced, it is transported into the cells by a Fe2+ transporter encoded by IRT1 in Arabidopsis [18] [3].
Under Fe deficiency conditions, dicotyledonous plants induce physiological and morphological changes in their roots, known as “Fe deficiency responses”, aimed to improve its mobilization and uptake. The main physiological responses are: enhanced ferric reductase activity, enhanced Fe2+ transport, and rhizosphere acidification, due to the upregulation of FERRIC REDUCTATE OXIDASE (FRO), IRON-REGULATED TRANSPORTER (IRT) and H+-ATPase (HA) genes [19,20-22][4][5][6][7]. Other relevant physiological responses are the increased synthesis and/or release of organic acids; phenolic compounds, such as coumarins; and flavins, which can act as chelating and reducing Fe agents, improving its solubility for plants [23-27] [8][9][10][11][12].
Morphological responses are aimed to increase the contact surface of roots with soil and include development of subapical root hairs, of cluster roots (also named proteoid roots) and of transfer cells [32,33][13][14]. Physiological responses are generally located in the subapical regions of roots [32][13].
The master regulator of most Fe deficiency responses in Arabidopsis is FIT, a basic helix-loop-helix (bHLH) transcription factor (TF) which is induced in Fe-deficient roots [34-36] [15][16][17].
Plants acquire P from the soil as inorganic phosphate (Pi) through phosphate transporters, encoded by PHOSPHATE TRANSPORTER 1 (PHT1)-like genes, which are induced by P deficiency [55] [18]. In Arabidopsis, the main Pi transporters implicated in its acquisition from the medium are PHT1;1 and PHT1;4, also named PT1 and PT2 [15,56-59] [19][20][21][22][23]. Both transporters are located in subapical root hairs [60,61][24][25].
When plants grow under P deficiency conditions, they induce several physiological and morphological responses, mainly in their roots, to improve its acquisition. The main physiological responses are the increase of phosphate transporters; the acidification of the rhizosphere; and the exudation to the rhizosphere of phosphatases, encoded by genes like PAP17 in Arabidopsis (also named ACP5, “Acid Phosphatase”), and organic acids [51,58,59,62][26][22][23][27].
Morphological responses to P deficiency include primary root growth inhibition, development of lateral roots, root hairs, transfer cells [72,80-82][28][29][30][31] and an increase of the root/shoot ratio [83,84][32][33]. Some plants species develop cluster roots when grown under P (also under Fe) deficiency conditions [63,64,72,82][34][35][28][31].
The regulation of phosphate transporter genes, like PHT1;1 and PHT1;4, is mediated by several TFs, such as the PHOSPHATE RESPONSE (PHR1) and the PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) [67][36]. PHR1 regulates the expression of about 60% of the P deficiency related genes [68] [37]. Several P deficiency related genes not regulated by PHR1 are regulated by PHL (PHR1-like) TFs [68] [37].
Finally, another important response to P deficiency is the internal phosphate remobilization, which includes Pi vacuolar release and membrane phospholipids change by sulpholipids and galactolipids [63,64,78] [37].
Fe/P interactions are, therefore, numerous and varied, and can determine the efficiency of Fe and P nutrition in plants. The control of the Fe/P interactions could be a new and effective way to improve the P and Fe nutrition of crops [91][38]. However, it is necessary to deeply know the mechanisms underpinning their interactions to ensure the success and to obtain more efficient genotypes in the acquisition of both nutrients.
2. Role of ET and NO in the Regulation of Fe and P Deficiency Responses. Similarities and Differences
Fe and P deficiency responses, which enable plants to improve their ability to acquire these nutrients, show many similarities [58,59,94][39][40][41]. Several responses to both deficiencies are similar, like rhizosphere acidification, enhanced synthesis of organic acids and coumarins, and development of subapical root hairs and proteoid roots. Furthermore, both kinds of responses share common regulators, such as ET and NOnitric oxide (NO), which increase their production in Fe-deficient roots as well as in P-deficient roots [33,81,82,95,96,97,98,99,100,101][33][42][43][44][45][46][47][48][49][50].
2.1. ET and NO Involvement in the Regulation of Physiological and Morphological Responses to Fe Deficiency
ET/NO have been involved in the activation of physiological responses to Fe deficiency and in most of the Fe-related genes associated with them [22,102,103,104,105,106,107][22][51][52][53][54][55][56]. ET/NO have also been involved, along with auxin and other signaling substances, in the development of most of the Fe deficiency morphological responses [reviewed in 33 and 81]. ET/NO interactions in such a regulation was studied by Romera’s group [107,108,109]. Furthermore, this group, along with Dr. Yeh’s group, has also paved the way to study the relationship between NO and S-nitrosoglutathione (GSNO) in the regulation of Fe deficiency responses [110,111,112]. NO can react with glutathione (GSH) to produce GSNO, the main NO reservoir in plants [113]. GSNO levels are regulated by the GSNOR enzyme [112]. The GSNOR1 gene, encoding this enzyme in Arabidopsis, is upregulated under Fe deficiency [112]. GSNO and NO have been found to play key roles in the regulation of Fe deficiency responses and other abiotic and biotic stress responses [34,106,107,108,110,111,112,114,115,116].
ET and NO have also been involved in the regulation of the cell wall composition and in the dynamic of the endodermal cells suberization, respectively. Fe deficiency increases the synthesis of putrescine, a natural polyamine that triggers a NO burst, which acts as a positive regulator of the solubilization of cell wall-bound Fe [117].
2.2. ET and NO Involvement in the Regulation of Physiological and Morphological Responses to P Deficiency
For decades, the ET involvement in the regulation of P deficiency responses has been limited to its role in the inhibition of the primary root growth and in the development of lateral roots and root hairs [96,118,119,120]. Nowadays, there are experimental results showing that ET also plays an important role in the regulation of physiological responses to P deficiency [57,99,121,122,123]. Lei et al. [121] showed that the ET insensitive Arabidopsis mutants etr1 and ein2-5 present a lower expression of the genes encoding the P transporters PT1(PHT1;1) and PT2(PHT1;4). On the contrary, Arabidopsis hps2 (ctr1 homolog), a constitutive ET signaling mutant [124], displays enhanced responses to P deficiency [121]. ET has also been involved in PHT1;5 regulation [57,79]. Finally, very recently it has been shown that PHR1 expression is induced by ACC (ET precursor) [69].
In other plant species, such as Medicago falcata, results also support a role for ET in the regulation of P deficiency responses [125]. The expression of MfPT1 and MfPT5, encoding P transporters, and of MfPAP1, encoding an acid phosphatase, was clearly inhibited by the ET synthesis inhibitors aminoethoxyvinyl glycine (AVG) and cobalt (Co2+). On the other hand, the treatment with the ET precursor ACC produced an increase of the expression of those genes in plants grown under P-sufficient conditions [125].
To date, the role of NO, GSNO and GSNOR in the regulation of physiological responses to P deficiency has been poorly studied [101]. However, in a recent work [126] it has been shown that NO induces the expression of the P transporter OsPT2. On the other hand, and as occurs under Fe deficiency, the GSNOR1 gene is also upregulated under P deficiency [112].
For many years, ET has been involved in the regulation of morphological responses to Fe and P deficiency, like subapical roots hairs, proteoid roots and transfer cells [reviewed in 33,81,104]. For example, Zaid et al. [95] found that proteoid root formation in Casuarina glauca, induced by Fe deficiency, was inhibited by treatments with several ET synthesis and action inhibitors, like aminooxyacetic acid (AOA), cobalt and silver thiosulfate (STS), while the ET precursor ACC induced the development of proteoid roots in Fe-sufficient plants. These results clearly show a role for ET in this typical morphological response to either Fe or P deficiency.
In addition to ET, NO has been involved in the regulation of morphological responses to Fe and P deficiency [100,101,106]. The development of proteoid roots in lupine plants growing under Fe and P sufficiency, and the expression of LaSCR1 and LaSCR2, two essential genes related to the development of proteoid roots, were induced by GSNO (NO donor) treatment [97,127]. On the other hand, the development of proteoid roots, and the expression of the genes previously mentioned, were inhibited in Fe- or P-deficient plants by treatment with cPTIO (2-4-carboxyphenyl-4, 4, 5, 5-tetramethylimidazoline-1-oxyl-3-oxide), a NO chelating agent [127].
3. Mechanisms for the induction of Fe- and P-related genes by ET and NO
The mechanisms by which ET and NO regulate the responses to both deficiencies are not completely known although some advances have been achieved in the last years. For ET, it has been found that EIN3/EIL1 are common key TFs involved in the regulation of physiological and morphological responses to Fe and P deficiency (Figure 1).
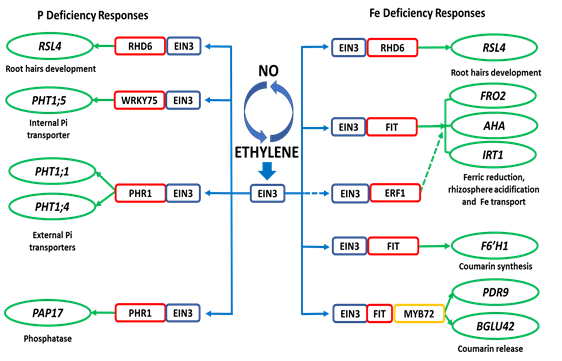
Figure 1. Working model proposed to explain the regulation of P and Fe deficiency responses by ET through EIN3. EIN3 (also EIL1) interacts with the PHR1 and WRKY75 TFs to regulate some P deficiency responses, such as expression of the internal (PHT1;5) and external (PHT1;1 and PHT1;4) Pi transporters, and PAP17 (acid phosphatase). EIN3 (also EIL1) interacts with the FIT TF to regulate some Fe deficiency responses, such as expression of FRO2 (ferric reductase), IRT1 (Fe transporter), F6´H1, BGLU42 and PDR9 (genes related to coumarin synthesis and release; these latter ones activated through the MYB72 TF). EIN3 can also interact with the ERF1 TF to regulate FRO2 and IRT1 expression. Finally, EIN3 can also interact with RHD6 to modify the expression of RSL4 and promotes root hair elongation either under P deficiency or Fe deficiency.
Based on, 33, 40, 41, 46, 69, 96, 99, 131, 135,136, 137[14][57][58][59][60][61][62][63][64][65][66].
In relation to the role of NO and GSNO in the regulation of different Fe acquisition processes, right now it is very difficult to discriminate between both compounds. However, in a recent work, by using a chemical screening approach, it has been proposed that NO and GSNO have different roles in the regulation of Fe acquisition genes [111][67].
The positive and reciprocal influence between NO and ET in the regulation of Fe and Mg deficiency responses is already known [108,109,138][68][69][70]. However, the interrelationship between GSNO and ET is also feasible. It seems that the main way for GSNO to regulate Fe deficiency responses is through the reversible S-nitrosylation of proteins [139-141][71][72][73]. Among them are some enzymes involved in ET synthesis, such as SAM synthetases [142][74], which can be inhibited by S-nitrosylation [143,144] [75][39]. In this way, higher GSNO levels (such as those found in Fe-sufficient roots) could contribute to S-nitrosylation of ET synthesis enzymes and, consequently, to the inhibition of ET synthesis [112][40], while lower GSNO levels (such as those found in Fe-deficient roots) could contribute to de-nitrosylation of ET synthesis enzymes and, consequently, to an increase of ET synthesis [112][40] (Figure 2).
Besides ET synthesis, GSNO (NO) could also affect ET signaling. Very recently, it has been found that the ERF72 TF (also named RAP2.3) participates in the regulation of Fe deficiency responses in Arabidopsis [132][76]. ERF72 belongs to the group VII ERFs [145][77], which are sensors of NO and can be targeted for proteolysis degradation by the N-end rule in the presence of NO [146][78]. Curiously, ERF72 interacts with DELLA proteins [145][77], also involved in the regulation of Fe deficiency responses [147][79].
GSNO/NO levels can be regulated by ET. In a recent work, García et al. [112][40] showed that ET (ACC) induces GSNOR1 expression (Figure 2). ET (ACC) can also increase NO content by activating enzymes involved in its synthesis, such as nitrate reductase and nitric oxide synthase-like [138][70]. This would imply that ET could simultaneously increase NO accumulation and decrease GSNO content [Figure 2; 112][40].
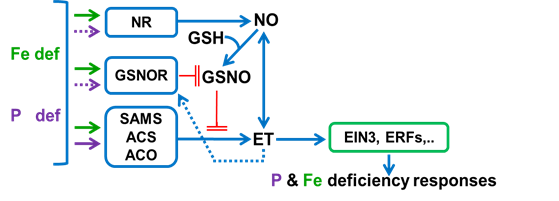
Figure 2. Model to summarize the relationship between ET, NO and GSNO in Fe- and P-deficient roots. Fe and P deficiency conditions induce several enzymes related to NO and ET synthesis, like NR, SAMS, ACS and ACO. Each one (NO, ET) mutually influences the synthesis of each other. Fe and P deficiency can also induce the GSNOR enzyme, which leads to a decrease in GSNO levels. GSNO, formed from NO and GSH, can limit ET synthesis through SAMS nitrosylation. On the other hand, ET can promote GSNO degradation by inducing the GSNOR enzyme. Finally, ET, through EIN3 and other ERF TFs, would activate Fe and P deficiency responses.
ET, ethylene; GSH, glutathione; GSNO, S-nitrosoglutathione; NO, nitric oxide. NR, nitrate reductase; GSNOR, GSNO reductase; SAMS, S-Adenosylmethionine synthetase; ACS, ACC syntase; ACO, ACC oxidase. à : promotion (dashed line indicates promotion based on preliminary results); ─╢: inhibition
Based on 112, 138, 142, 143, 144[40][70][74][75][39].
Although NO has also been studied in relation to P deficiency morphological responses [97,127], the role of endogenous GSNO and GSNOR in the regulation of P deficiency responses has hardly been studied [101].
References
- Lucena, C.; Porras, R.; Romera, F.J.; Alcántara, E.; García, M.J.; Pérez-Vicente, R. Similarities and Differences in the Acquisition of Fe and P by Dicot Plants. Agronomy 2018, 8, 148.
- Lucena, C.; Porras, R.; García, M.J.; Alcántara, E.; Pérez-Vicente, R.; Zamarreño-Ángel, M.; Bacaicoa, E.; García-Mina, J.M.; Smith, A.P.; Romera, F.J. Ethylene and Phloem Signals Are Involved in the Regulation of Responses to Fe and P Deficiencies in Roots of Strategy I Plants. Front. Plant. Sci. 2019, 10, 1237.
- Venuti, S.; Zanin, L.; Marroni, F.; Franco, A.; Morgante, M.; Pinton, R.; Tomasi, N. Physiological and transcriptomic data highlight common features between iron and phosphorus acquisition mechanisms in white lupin roots. Plant. Sci. 2019, 285, 110–121.
- Lucena, C.; Romera, F.J.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene Participates in the Regulation of Fe Deficiency Responses in Strategy I Plants and in Rice. Front. Plant. Sci. 2015, 6, 1056.
- García, M.J.; Romera, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R. Ethylene and the Regulation of Physiological and Morphological Responses to Nutrient Deficiencies. Plant. Physiol. 2015, 169, 51–60.
- Neumann, G. The Role of Ethylene in Plant Adaptations for Phosphate Acquisition in Soils—A Review. Front. Plant. Sci. 2016, 6, 1224.
- Zaid, H.; El Morabet, R.; Diem, H.G.; Arahou, M. Does ethylene mediate cluster root formation under iron deficiency? Ann. Bot. 2003, 92, 673–677.
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The Molecular Mechanism of Ethylene-Mediated Root Hair Development Induced by Phosphate Starvation. PLoS Genet. 2016, 12, e1006194.
- Wang, B.L.; Tang, X.Y.; Cheng, L.Y.; Zhang, A.Z.; Zhang, W.H.; Zhang, F.S.; Liu, J.Q.; Cao, Y.; Allan, D.L.; Vance, C.P.; et al. Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol. 2010, 187, 1112–1123.
- García, M.J.; Angulo, M.; García, C.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R.; Romera, F.J. Influence of Ethylene Signaling in the Crosstalk Between Fe, S, and P Deficiency Responses in Arabidopsis thaliana. Front. Plant. Sci. 2021, 12, 643585.
- Song, L.; Liu, D. Ethylene and plant responses to phosphate deficiency. Front. Plant. Sci. 2015, 6, 796.
- Buet, A.; Galatro, A.; Ramos-Artuso, F.; Simontacchi, M. Nitric oxide and plant mineral nutrition: Current knowledge. J. Exp. Bot. 2019, 70, 4461–4476.
- Galatro, A.; Ramos-Artuso, F.; Luquet, M.; Buet, A.; Simontacchi, M. An Update on Nitric Oxide Production and Role Under Phosphorus Scarcity in Plants. Front. Plant. Sci. 2020, 11, 413.
- Waters, B.M.; Lucena, C.; Romera, F.J.; Jester, G.G.; Wynn, A.N.; Rojas, C.L.; Alcántara, E.; Pérez-Vicente, R. Ethylene involvement in the regulation of the H+-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron trasnporter CsIRT1 genes in cucumber plants. Plant Physiol. Biochem. 2007, 45, 293–301.
- Romera, F.J.; Alcantara, E. Iron-Deficiency Stress Responses in Cucumber (Cucumis sativus L.) Roots (A Possible Role for Ethylene?). Plant. Physiol. 1994, 105, 1133–1138.
- Romera, F.J.; Alcantara, E.; de la Guardia, M.D. Ethylene Production by Fe-deficient Roots and its Involvement in the Regulation of Fe-deficiency Stress Responses by Strategy I Plants. Ann. Bot. 1999, 83, 51–55.
- Romera, F.J.; Alcántara, E. Ethylene involvement in the regulation of Fe-deficiency stress responses by Strategy I plants. Funct. Plant. Biol. 2004, 31, 315–328.
- Lucena, C.; Waters, B.M.; Romera, F.J.; García, M.J.; Morales, M.; Alcántara, E.; Pérez-Vicente, R. Ethylene could influence ferric reductase, iron transporter and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J. Exp. Bot. 2006, 57, 4145–4154.
- Graziano, M.; LaMattina, L. Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant. J. 2007, 52, 949–960.
- García, M.J.; Lucena, C.; Romera, F.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 2010, 61, 3885–3899.
- Nagarajan, V.K.; Smith, A.P. Ethylene’s Role in Phosphate Starvation Signaling: More than Just a Root Growth Regulator. Plant. Cell Physiol. 2011, 53, 277–286.
- Lucena, C.; Porras, R.; Romera, F.J.; Alcántara, E.; García, M.J.; Pérez-Vicente, R. Similarities and Differences in the Acquisition of Fe and P by Dicot Plants. Agronomy 2018, 8, 148.
- Lucena, C.; Porras, R.; García, M.J.; Alcántara, E.; Pérez-Vicente, R.; Zamarreño-Ángel, M.; Bacaicoa, E.; García-Mina, J.M.; Smith, A.P.; Romera, F.J. Ethylene and Phloem Signals Are Involved in the Regulation of Responses to Fe and P Deficiencies in Roots of Strategy I Plants. Front. Plant. Sci. 2019, 10, 1237.
- Karthikeyan, A.S.; Varadarajan, D.K.; Mukatira, U.T.; D’Urzo, M.P.; Damsz, B.; Raghothama, K.G. Regulated Expression of Arabidopsis Phosphate Transporters. Plant. Physiol. 2002, 130, 221–233.
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant. J. 2002, 31, 341–353.
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant. Biol. 2014, 65, 95–123.
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741.
- Wang, Z.; Straub, D.; Yang, H.; Kania, A.; Shen, J.; Ludewig, U.; Neumann, G. The regulatory network of cluster-root function and development in phosphate-deficient white lupin (Lupinus albus) identified by transcriptome sequencing. Physiol. Plant. 2014, 151, 323–338.
- Lynch, J.P.; Brown, K.M. Topsoil foraging—An architectural adaptation of plants to low phosphorus availability. Plant. Soil 2001, 237, 225–237.
- García, M.J.; Romera, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R. Ethylene and the Regulation of Physiological and Morphological Responses to Nutrient Deficiencies. Plant. Physiol. 2015, 169, 51–60.
- Neumann, G. The Role of Ethylene in Plant Adaptations for Phosphate Acquisition in Soils—A Review. Front. Plant. Sci. 2016, 6, 1224.
- Lynch, J. Root Architecture and Plant Productivity. Plant. Physiol. 1995, 109, 7–13.
- Lynch, J.; Brown, K.M. Ethylene and plant responses to nutritional stress. Physiol. Plant. 1997, 100, 613–619.
- Cheng, L.; Bucciarelli, B.; Liu, J.; Zinn, K.; Miller, S.; Patton-Vogt, J.; Allan, D.; Shen, J.; Vance, C.P. White Lupin Cluster Root Acclimation to Phosphorus Deficiency and Root Hair Development Involve Unique Glycerophosphodiester Phosphodiesterases. Plant. Physiol. 2011, 156, 1131–1148.
- Cheng, L.; Bucciarelli, B.; Shen, J.; Allan, D.; Vance, C.P. Update on White Lupin Cluster Root Acclimation to Phosphorus Deficiency Update on Lupin Cluster Roots. Plant. Physiol. 2011, 156, 1025–1032.
- Chen, J.; Liu, Y.; Ni, J.; Wang, Y.; Bai, Y.; Shi, J.; Gan, J.; Wu, Z.; Wu, P. OsPHF1 Regulates the Plasma Membrane Localization of Low- and High-Affinity Inorganic Phosphate Transporters and Determines Inorganic Phosphate Uptake and Translocation in Rice. Plant. Physiol. 2011, 157, 269–278.
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102.
- Ward, J.T.; Lahner, B.; Yakubova, E.; Salt, D.E.; Raghothama, K.G. The Effect of Iron on the Primary Root Elongation of Arabidopsis during Phosphate Deficiency. Plant. Physiol. 2008, 147, 1181–1191.
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH Transcription Factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 Reveals Molecular Linkage between the Regulation of Iron Acquisition and Ethylene Signaling in Arabidopsis. Plant. Cell 2011, 23, 1815–1829.
- Yang, Y.; Ou, B.; Zhang, J.; Si, W.; Gu, H.; Qin, G.; Qu, L.-J. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant. J. 2014, 77, 838–851.
- Curie, C.; Mari, S. New routes for plant iron mining. New Phytol. 2017, 214, 521–525.
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and Ethylene Coordinately Regulate the Phosphate Starvation Response through Transcriptional Regulation of PHOSPHATE STARVATION RESPONSE1. Plant. Cell 2017, 29, 2269–2284.
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The Molecular Mechanism of Ethylene-Mediated Root Hair Development Induced by Phosphate Starvation. PLoS Genet. 2016, 12, e1006194.
- Song, L.; Liu, D. Ethylene and plant responses to phosphate deficiency. Front. Plant. Sci. 2015, 6, 796.
- Balparda, M.; Armas, A.M.; Estavillo, G.M.; Roschzttardtz, H.; Pagani, M.A.; Gomez-Casati, D.F. The PAP/SAL1 retrograde signaling pathway is involved in iron homeostasis. Plant. Mol. Biol. 2020, 102, 323–337.
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A Tripartite Amplification Loop Involving the Transcription Factor WRKY75, Salicylic Acid, and Reactive Oxygen Species Accelerates Leaf Senescence. Plant. Cell 2017, 29, 2854–2870.
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839.
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133.
- Kailasam, S.; Wang, Y.; Lo, J.-C.; Chang, H.-F.; Yeh, K.-C. S-Nitrosoglutathione works downstream of nitric oxide to mediate iron-deficiency signaling in Arabidopsis. Plant. J. 2018, 94, 157–168.
- García, M.J.; Suárez, V.; Romera, F.J.; Alcántara, E.; Pérez-Vicente, R. A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant. Physiol. Biochem. 2011, 49, 537–544.
- Plant Signaling & Behavior. Plant. Signal. Behav. 2015, 6, 167–170.
- Liu, M.; Zhang, H.; Fang, X.; Zhang, Y.; Jin, C. Auxin Acts Downstream of Ethylene and Nitric Oxide to Regulate Magnesium Deficiency-Induced Root Hair Development in Arabidopsis thaliana. Plant. Cell Physiol. 2018, 2018, 591452.
- Malik, S.I.; Hussain, A.; Yun, B.-W.; Spoel, S.H.; Loake, G.J. GSNOR-mediated de-nitrosylation in the plant defence response. Plant. Sci. 2011, 181, 540–544.
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Mata-Pérez, C.; Valderrama, R.; Padilla, M.N.; López-Jaramillo, J.; Luque, F.; Corpas, F.J.; Barroso, J.B. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J. Exp. Bot. 2015, 66, 5983–5996.
- Zaffagnini, M.; de Mia, M.; Morisse, S.; di Giacinto, N.; Marchand, C.; Maes, A.; Lemaire, S.; Trost, P. Protein S-nitrosylation in photosynthetic organisms: A comprehensive overview with future perspectives. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 952–966.
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154.
- Lindermayr, C.; Saalbach, G.; Bahnweg, G.; Durner, J. Differential Inhibition of Arabidopsis Methionine Adenosyl transferases by Protein S-Nitrosylation. J. Biol. Chem. 2006, 281, 4285–4291.
- Efreschi, L. Nitric oxide and phytohormone interactions: Current status and perspectives. Front. Plant. Sci. 2013, 4, 398.
- García, M.J.; Corpas, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R.; Zamarreño, Á.M.; Bacaicoa, E.; García-Mina, J.M.; Bauer, P.; Romera, F.J. A Shoot Fe Signaling Pathway Requiring the OPT3 Transporter Controls GSNO Reductase and Ethylene in Arabidopsis thaliana Roots. Front. Plant. Sci. 2018, 9, 1325.
- Liu, W.; Li, Q.; Wang, Y.; Wu, T.; Yang, Y.; Zhang, X.; Han, Z.; Xu, X. Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem. Biophys. Res. Commun. 2017, 491, 862–868.
- La Rosa, N.M.-D.; Sotillo, B.; Miskolczi, P.; Gibbs, D.J.; Vicente, J.; Carbonero, P.; Oñate-Sánchez, L.; Holdsworth, M.J.; Bhalerao, R.; Alabadí, D.; et al. Large-Scale Identification of Gibberellin-Related Transcription Factors Defines Group VII ETHYLENE RESPONSE FACTORS as Functional DELLA Partners. Plant. Physiol. 2014, 166, 1022–1032.
- Gibbs, D.J.; Isa, N.M.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; la Rosa, N.M.-D.; Conde, J.V.; Correia, C.S.; Pearce, S.P.; et al. Nitric Oxide Sensing in Plants Is Mediated by Proteolytic Control of Group VII ERF Transcription Factors. Mol. Cell 2014, 53, 369–379.
- Wild, M.; Davière, J.-M.; Regnault, T.; Sakvarelidze-Achard, L.; Carrera, E.; Diaz, I.L.; Cayrel, A.; Dubeaux, G.; Vert, G.; Achard, P. Tissue-Specific Regulation of Gibberellin Signaling Fine-Tunes Arabidopsis Iron-Deficiency Responses. Dev. Cell 2016, 37, 190–200.
