Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Christiane Funk and Version 2 by Lily Guo.
FtsH metalloproteases found in eubacteria, animals, and plants are well-known for their vital role in the maintenance and proteolysis of membrane proteins. Their location is restricted to organelles of endosymbiotic origin, the chloroplasts, and mitochondria. In the model organism Arabidopsis thaliana, there are 17 membrane-bound FtsH proteases containing an AAA+ (ATPase associated with various cellular activities) and a Zn2+ metalloprotease domain.
- AAA-type protease
- FtsH metalloprotease
- chloroplast
- embryo lethal
- leaf variegation
- plastid biogenesis
- protein import
- oxidative stress
1. Introduction
Cells have evolved an extensive system of molecular chaperones, folding catalysts, and proteases that control protein quality and prevent damage. In addition to the well-studied degradative removal of damaged or superfluous proteins [1], proteolysis is highly important in regulating protein preprocessing, maturation, post-translational protein modifications, and signaling [2][3][2,3]. Therefore, it is is no overstatement that proteolysis is directly or indirectly involved in most cellular processes [4].
1.1. Proteases in the Plant Chloroplast
Plant proteases associated with a particular proteolytic activity are present in various cellular compartments and organelles constituting up to 3% of the plant proteome [5][6][5,6]. Proteases are classified according to their catalytic types. Except for glutamic acid proteases, representatives from all protease classes (threonine, cysteine, serine, aspartic, metalloproteases) have been detected in the plant Arabidopsis thaliana [7]. The chloroplast is a unique organelle of the plant cell; absorption and conversion of light energy in the photosynthetic reaction lead to a permanent need for protein turnover (processing and degradation) to adapt to different light conditions. Excess light adsorption further can cause the formation of reactive oxygen species and damage proteins. Therefore, protein quality and quantity controls are essential [8][9][8,9]. In addition to photosynthesis, several metabolic reactions happen in the chloroplast, including the biosynthesis of lipids, amino acids, chlorophylls, and carotenoids; therefore, plastidic proteases are vital regulators [9]. More than 20 different families of chloroplast proteases have been detected, with members localized in specific sub-organellar compartments [3][9][3,9].
1.2. Plant Pseudo-Proteases
In addition to the proteolytically active proteases, members with mutations in their active site attracted the attention of researchers. Despite their putative proteolytically inactivity, many of these pseudo-proteases have essential roles in the cell, ranging from structural proteins via chaperones [10][11][10,11] to enzymes with a new function [12][13][14][15][16][17][18][19][20][12,13,14,15,16,17,18,19,20]. In the chloroplast, pseudo-proteases are found in the families of serine Clp-proteases (ClpRs, [21][22][23][21,22,23]) and FtsH metalloproteases (FtsHis [18][24][25][26][18,24,25,26]). ClpRs of the Clp protease family lack their catalytic triad. These proteolytically inactive subunits are of structural importance to form a tetradecameric proteolytic core together with the catalytically active ClpP subunits [21][22][23][21,22,23]. Most ClpR proteins are essential for the proteolytic function of the Clp core function.
This review will focus on pseudo-proteases belonging to the family of the membrane-bound ATP-dependent FtsH metalloproteases, which are termed FtsHi (i-inactive) [23]. These presumably proteolytically inactive FtsHi enzymes are restricted to the plant chloroplast and, similar to the ClpR, affect chloroplast and overall plant development [17][23][24][25][17,23,24,25].
2. Filamentation Temperature-Sensitive Protein H (FtsH) Protease Family
The name FtsH (filamentation temperature-sensitive) erroneously originated from the growth behavior of a Y16 Escherichia coli strain deficient in its ftsh gene. However, later, a second, independent mutation was found to be responsible for the observed temperature-sensitive phenotype [23].
FtsH proteases, known as zincins, belong to the MEROPS peptidase family M41, which in turn belongs to a larger family of zinc metalloproteases [27]. Within the M41 peptidase domain, the Zn2+ ion is ligated by two histidine residues, forming the HEXXH motif (where X is any uncharged residue) as well as a glutamic acid residue [28]. Functional homo- or hetero-hexameric complexes are inserted into the membrane by one or more N-terminal transmembrane domains per subunit [29][30][31][32][29,30,31,32]. The highly conserved AAA+ domain, a cassette of about 200–250 residues that contains the ATP-binding motif (Walker A and Walker B) and the second region of homology (SRH), is situated between the transmembrane region and the active site (Figure 1). Unlike other well-studied ATP-dependent proteases, FtsHs lack robust unfoldase activity [33]. Instead, ATP hydrolysis by FtsH is used to translocate unfolded substrates sequentially into the hexameric pore [33][34][33,34]. The AAA+ domain is required for nucleotide binding and hydrolysis [29][35][29,35] and responsible for alternating between a closed and open state of the FGV pore motif, which is a conserved hydrophobic area at the proteolytic chamber [29]. The substrate is pulled into the degradation chamber via a narrow pore [36][37][36,37]. Recent cryo-electron microscopy studies enabled the study of substrate processing of AAA+ proteins in detail (reviewed in [38]) and revealed a conserved spiraling organization of ATPase hexamers around the translocating protein substrate.
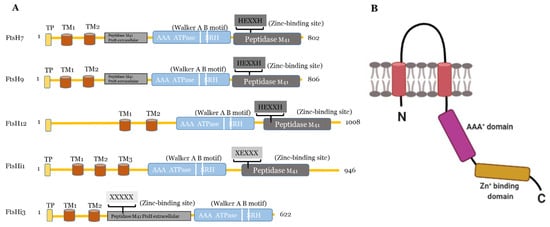
Figure 1.
(
A
) Predicted domains and motifs of AtFtsHis in comparison to the presumably active AtFtsH7, 9, and 12. TP, transit peptide; transmembrane domains, TM1-3; Walker A B motifs are indicated as two white lines between the AAA
+
ATPase and SRH; SRH, second region of homology. Active FtsH proteases contain the Zn
2+
-binding motif (HEXXH) in the their peptidase M41 domain, which is substituted or absent in presumably inactive FtsHis (substitution of both histidines indicated as XEXXX). In FtsHi3, the peptidase M41 domain is annotated as “FtsH extracellular” and is located at the N-terminal to the ATPase; its HEXXH motif is completely missing (XXXXX). FtsH7 and FtsH9 contain an FtsH extracellular” peptidase domain additionally to their protease domain. AtFtsHis are predicted to contain three (FtsHi1, 5), two (FtsHi2, 3), or one (FtsHi4) transmembrane domains (
B
) Schematic drawing of the structure of a monomeric FtsH protease with two membrane-spanning regions (shown in red), the proteolytic domain (in range) and the AAA
+
-domain (in pink). Created with BioRender.com (accessed on 27 May 2021).
2.1. The FtsH Protease Family of Arabidopsis thaliana
The annual plant Arabidopsis thaliana contains 17 different FtsH proteases. Gene comparison studies showed that of the 12 FTSH genes potentially coding for fully functional proteases, ten are found in highly homologous pairs. While the pairs AtFtsH1/5, AtFtsH2/8, and AtFtsH7/9 are targeted to the chloroplast, AtFtsH3/10 and AtFtsH4 have been identified in mitochondria. AtFtsH11, the pair partner of AtFtsH4, was initially reported to be dual targeted to mitochondria and the chloroplast [39]. However, Wagner and coworkers confirmed its location to be only in chloroplasts [40]. AtFtsH3 and AtFtsH10 were shown not to be crucial for growth under optimal conditions [41]. Loss of AtFtsH4 leads to oxidative stress and the accumulation of oxidized proteins [42][43][42,43]. FtsH10 is involved in the assembly and/or stability of complexes I and V of the mitochondrial oxidative phosphorylation system [43].
2.1.1. FtsH Proteases Located in the Thylakoid Membrane
Of the plastidic FtsHs, FtsH1, 2, 5, and 8 are localized in the thylakoid membrane. These members are the most abundant FtsH proteases and extensively studied [44]; they form hetero-hexameric complexes, in which FtsH1 and FtsH5 (Type A) and FtsH2 and FtsH8 (Type B) can partially substitute for each other [45]. A threshold in the amount of type A and B subunits was postulated to determine the proper function and development of chloroplasts and thylakoid membrane [46][47][48][49][46,47,48,49]. This thylakoid located protease complex plays a vital role in the degradation and assembly of the Photosystem II reaction center protein D1 and other transmembrane subunits of the photosynthetic machinery. Mutants lacking FtsH2/VAR2 or FtsH5/VAR1 show strongly or slightly variegated leaves, respectively [50][51][50,51]. Functional loss of, e.g., FtsH2 results in upregulation of other FtsH proteins in the green leaf sectors to maintain proper function and development of the chloroplasts [47][50][52][47,50,52]. FtsH6 is also localized in the thylakoid membrane. It is essential for thermotolerance and thermomemory in seedlings [53], while no phenotype was observed in adult plants when grown in semi-natural outdoor conditions [54].
2.1.2. FtsH Proteases Located in the Chloroplast Envelope
The other plastidic FtsH enzymes are believed to be localized in the chloroplast envelope [19]. Deleting FtsH7 and 9 does not result in any obvious phenotype [53], and the proteases are not required for PSII repair [55]. FtsH11 is crucial for growth in long photoperiods [40] and thermotolerance [56][57][56,57]. FtsH12 was co-immuno-precipitated in a complex with FtsHi1, 2, 4, 5 and NAD-dependent malate dehydrogenase (MDH) and shown to be involved in protein import [58][59][58,59]. In addition to FtsHi1, 2, 4, and 5, even FtsHi3 belongs to the five plastidic FtsH homologues, which are incapable of proteolysis in Arabidopsis. The FtsHi enzymes either have a mutation in their HEXXH motif (FtsHi1, 2, 4, and 5), or the entire motif is missing (FtsHi3) [18][26][18,26]. Compared to AtFtsHi1, 2, 4, and 5, FtsHi3 contains a very short C-terminal domain. Interestingly, FtsHi3 also has undergone a domain swap: the whole M41 domain is located at the N-terminal instead of at the C-terminal to the AAA+ domain [26]. Comparing the domain organization of AtFtsHi3 with AtFtsHi1, AtFtsH7, 9, 12 (Figure 1), also AtFtsH7/9 contain this “peptidase M41 FtsH extracellular” domain N-terminal to the AAA+ domain, which is additional to their protease domain located in the C-terminal to the AAA+ domain. This additional domain is not present in other FtsHs or FtsHis. Whether the N-terminal “peptidase M41 FtsH extracellular” domain of FtsH7, FtsH9, and FtsHi3 enables these enzymes to form a common complex with a specific function remains to be shown. Three independent pre-protein translocating models (pSSU-TEV-protein A, pL11Flag-TEV-Protein A, pLHCP-TEV-protein A) suggested FtsHi3 to form a 1-MD complex separate from the FtsH12/FtsHi1,2,4,5/MDH complex [58] and different to the 1-MD TIC complex [60]. The identity of other components in this complex is unknown. FTSHi3 is not co-expressed with the tight cluster of FTSH12/FTSHi1, 2, 4, 5, but instead with a gene encoding OTP51 [26][58][59][26,58,59]. This pentatricopeptide repeat protein is required for the splicing of group IIa introns and impacts photosystem I and II assembly [61]. The tight co-expression with FTSHi3 indicates a common function of OTP51 and FtsHi3; therefore, OTP51 is another hypothetical complex partner.
2.1.2. FtsH Proteases Located in the Chloroplast Envelope
The other plastidic FtsH enzymes are believed to be localized in the chloroplast envelope [19]. Deleting FtsH7 and 9 does not result in any obvious phenotype [53], and the proteases are not required for PSII repair [55]. FtsH11 is crucial for growth in long photoperiods [40] and thermotolerance [56][57][56,57]. FtsH12 was co-immuno-precipitated in a complex with FtsHi1, 2, 4, 5 and NAD-dependent malate dehydrogenase (MDH) and shown to be involved in protein import [58][59][58,59]. In addition to FtsHi1, 2, 4, and 5, even FtsHi3 belongs to the five plastidic FtsH homologues, which are incapable of proteolysis in Arabidopsis. The FtsHi enzymes either have a mutation in their HEXXH motif (FtsHi1, 2, 4, and 5), or the entire motif is missing (FtsHi3) [18][26][18,26]. Compared to AtFtsHi1, 2, 4, and 5, FtsHi3 contains a very short C-terminal domain. Interestingly, FtsHi3 also has undergone a domain swap: the whole M41 domain is located at the N-terminal instead of at the C-terminal to the AAA+ domain [26]. Comparing the domain organization of AtFtsHi3 with AtFtsHi1, AtFtsH7, 9, 12 (Figure 1), also AtFtsH7/9 contain this “peptidase M41 FtsH extracellular” domain N-terminal to the AAA+ domain, which is additional to their protease domain located in the C-terminal to the AAA+ domain. This additional domain is not present in other FtsHs or FtsHis. Whether the N-terminal “peptidase M41 FtsH extracellular” domain of FtsH7, FtsH9, and FtsHi3 enables these enzymes to form a common complex with a specific function remains to be shown. Three independent pre-protein translocating models (pSSU-TEV-protein A, pL11Flag-TEV-Protein A, pLHCP-TEV-protein A) suggested FtsHi3 to form a 1-MD complex separate from the FtsH12/FtsHi1,2,4,5/MDH complex [58] and different to the 1-MD TIC complex [60]. The identity of other components in this complex is unknown. FTSHi3 is not co-expressed with the tight cluster of FTSH12/FTSHi1, 2, 4, 5, but instead with a gene encoding OTP51 [26][58][59][26,58,59]. This pentatricopeptide repeat protein is required for the splicing of group IIa introns and impacts photosystem I and II assembly [61]. The tight co-expression with FTSHi3 indicates a common function of OTP51 and FtsHi3; therefore, OTP51 is another hypothetical complex partner.
3. Pseudo-Proteases with an Important Enzymatic Activity
In addition to being pseudo-proteases, the AAA+ domain of FtsHis is intact and—based on the seed lethality of many FtsHi mutants—highly important for their activity, plastid, and overall plant development. Four out of the five AtFtsHi members have been demonstrated to form an inner envelope-bound heteromeric AAA+ (ATPase associated with diverse cellular activities) ATPase complex. This complex consisting of FtsH12/FtsHi1,2,4,5/pdNAD-MDH was found to be involved in ATP-driven protein import across the chloroplast envelope [58][59][62][63][58,59,62,63]. Even Ycf2 was observed being part of this 2 MDa complex using transgenic lines overexpressing FTSH12 [58]. Still, the protein could not be detected in complexes isolated from wild-type and tic56-3 plastids using a combination of native gel electrophoresis and protein quantification [64] and neither after pull-down of FtsH12 using its native promoter [65]. Kikuchi and coworkers [57] determined the super-complex Ycf2-FtsH12-FtsHi1,2,4,5-pdNAD-MDH physically to interact with TIC components such as Tic214 (Ycf1), Tic100, and Tic56 as well as with the pre-protein translocation components Toc75 and Toc159 [59]. In this complex, neither pdNAD-MDH activity [59] nor FtsH12 proteolytic activity [57] are required; an FtsH12 (H769Y) mutant developed normal chloroplasts with functional pre-protein import abilities.
The finding of ATP-driven protein import across the chloroplast membrane by an FtsH12/FtsHi1,2,4,5/pdNAD-MDH complex has steered an intense debate [66][67][66,67] on the importance of the long-accepted chloroplast protein import machinery, i.e., Tic110 and Tic40 forming a common translocon in the inner chloroplast membrane and recruiting the stromal chaperones Hsp93/ClpC1, cpHsp70, and Hsp90C [67]. If indeed the FtsH12/FtsHi complex plays the main role in protein import into the chloroplast [67] remains to be shown. The absence of this complex in most monocots (see Section 4) as well as its lower impact in adult plants (see Section 5) rather point to a specific function during chloroplast development in dicotyledons. We refer th
