Melanogenesis is the process leading to the synthesis of melanin, the main substance that influences skin color and plays a pivotal role against UV damage. Altered melanogenesis is observed in several pigmentation disorders. Melanogenesis occurs in specialized cells called melanocytes, physically and functionally related by means of autocrine and paracrine interplay to other skin cell types. Several external and internal factors control melanin biosynthesis and operate through different intracellular signaling pathways, which finally leads to the regulation of microphthalmia-associated transcription factor (MITF), the key transcription factor involved in melanogenesis and the expression of the main melanogenic enzymes, including TYR, TYRP-1, and TYRP-2. Epigenetic factors, including microRNAs (miRNAs), are involved in melanogenesis regulation. miRNAs are small, single-stranded, non-coding RNAs, of approximately 22 nucleotides in length, which control cell behavior by regulating gene expression, mainly by binding the 3′ untranslated region (3′-UTR) of target mRNAs.
- melanocyte
- melanogenesis
- microRNA
- skin pigmentation
1. Introduction
Skin represents the primary line of defense against environmental stressors, including chemical stimuli, microbial insults, allergens, and ultraviolet (UV) radiation. Protection from UV rays is essentially based on melanogenesis, the process leading to the synthesis of pigments called melanin, the main substance that influences skin color. Melanin protects the skin from harmful UV rays, as it can absorb UV and visible light and shows antioxidative and radical scavenging abilities, limiting UV-induced effects on cellular macromolecules, mainly DNA, thus protecting cells from genotoxic damage [1]. Therefore, reduced melanogenesis is also a major risk factor for melanoma and other skin cancers [2][3]. Nevertheless, increased melanogenesis and melanin accumulation is associated with hyperpigmentation disorders [4][5]. Skin hyperpigmentation, often associated with aging, hormonal changes, and UVB, is very common in clinical dermatology and includes dermal conditions such as melasma, chloasma, freckles, age spots, and sunspots [5]. In addition, both hyperpigmentation and hypopigmentation, as observed in vitiligo lesions, are frequently the consequence of inflammation, induced by skin stressors [6]. Indeed, the modulation of skin pigmentation still represents a challenge in treating dermatological disorders, despite several studies having investigated potential cures [7][8]. Melanin is produced by highly specialized cells called melanocytes that are in strict contact with other skin cells, especially keratinocytes. The process of skin coloration consists of melanin biosynthesis and the translocation of melanosomes, small organelles containing melanin, from melanocytes into epidermal keratinocytes [6][7][9].
Melanogenesis is a very complex process which includes the development, survival, and differentiation of melanocytes. It involves more than 150 genes and several signaling pathways that operate both at transcriptional and post-transcriptional levels in regulating the main melanogenic molecular players. These include transcription factors, enzymes, and regulatory molecules produced by melanocytes, as well as other skin cells including keratinocytes, dermal fibroblasts, and inflammatory and endocrine cells [6][7][10][11].
Altered gene expression and melanogenic regulatory factor activities are involved in melanogenesis dysfunction [4][11]. Increasingly, studies indicate that gene expression is also influenced by several epigenetic events, including chromatin modification, DNA methylation, and non-coding RNA classes such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) [12][13][14][15]. Indeed, the role of miRNAs in melanogenesis has been widely investigated.
2. Melanin Biosynthesis
Melanin synthesis occurs in highly specialized cells called melanocytes, localized in the basal layer of the epidermis and hair follicles [16][17]. Melanocytes consist of several ramifications called dendrites that end in keratinocytes, and each melanocyte is strictly connected to more than 30 keratinocytes, constituting the melanogenic unit [18]. During melanogenesis, a series of sequential reactions synthesize melanin, which is translocated into neighboring keratinocytes by means of melanosomes [6][7][9].
Melanin production is controlled by several enzymes including tyrosinase (TYR), tyrosine hydroxylase I (THI), and phenylalanine hydroxylase (PAH) in the initiation phase of melanin synthesis. Tyrosinase-associated protein 1 (TYRP-1) and tyrosinase-associated protein 2 (TYRP-2), also called dopachrome tautomerase (DCT), operate in the later phase [4][19]. TYR is a membrane-bound glycoprotein which plays a key role in the process, as it is considered the rate-limiting enzyme for melanin biosynthesis (
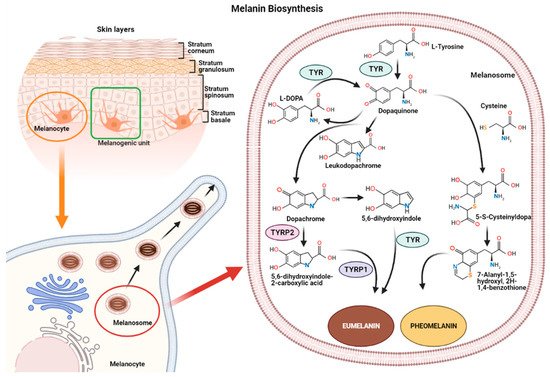
Figure 1.
left
right
The reaction catalyzed by TYR leads to L-tyrosine being transformed into dopaquinone by oxidation [20]. Dopaquinone is highly reactive and can follow two reaction chains from which eumelanin and pheomelanin originate:
- (1)
-
(1) In reactions leading to eumelanin production, dopaquinone undergoes intramolecular cyclization to produce leukodopachrome (cyclodopa). Cyclodopa undergoes redox exchange with another molecule of dopaquinone to form dopachrome and DOPA [20][21]. The dopachrome downstream process is branched in two ways. The first leads to the formation of 5,6-dihydroxyindole-2-carboxylic acid (DHICA) through TYRP-2 intervention and then into eumelanin by TYRP-1 conversion. The second leads to the conversion of dopachrome into 5,6-dihydroxyindole (DHI) and then into eumelanin involving TYR. At the end of this reaction, black-brownish eumelanin is formed.
- (2)
-
(2) In reactions leading to pheomelanin production, in the presence of cystein or glutathione, dopaquinone can be converted into 5-S-cysteinyldopa, or glutathionyldopa, which is then converted into quinoline and finally polymerized into red-yellow pheomelanin.In the skin, both eumelanin and pheomelanin form complex heteropolymers. The total amount of melanin, as well as the ratio between eumelanin to pheomelanin, are considered to determine skin color. Indeed, the role of the ratio between eumelanin and pheomelanin is still under debate, as many authors indicate that it remains unchanged in various phototypes of
human skin and, thus, skin color is dependent on total melanin amount [4][21]. Synthesized melanin is collected into melanosomes, which follow a complex maturation process and are transported to keratinocytes along actin filaments in association with motor proteins [4][22].
skin and, thus, skin color is dependent on total melanin amount [4,22]. Synthesized melanin is collected into melanosomes, which follow a complex maturation process and are transported to keratinocytes along actin filaments in association with motor proteins [4,23].3. Melanogenesis Regulation
Several stimuli control melanogenesis, including external factors such as UV rays or environmental pollution [11]. Furthermore, a series of endogenous molecules released by melanocytes, keratinocytes, dermal fibroblasts, and immune cells modulate the process paracrinally and autocrinally. These are mainly the α-melanocyte stimulating hormone (α-MSH) and the adrenocorticotropic hormone (ACTH), derived from the cleavage of pro-opiomelanocortin (POMC), from both melanocytes and keratinocytes [11][23]. In addition, stem cell factor (SCF), peptide endothelin 1 (ET-1), hepatocyte growth factor (HGF), keratinocyte growth factor (KGF), basic fibroblast growth factor (bFGF), and inflammatory mediators such as cytokines, prostaglandin E2 (PGE2), and nitric oxide (NO) significantly regulate melanogenesis [24][25]. All these stimuli operate with the activation of several signaling pathways [9][26]. Detailed descriptions of the multiple pathways in melanogenesis have been recently reported by others [6][7][9][11]. Here, the main signaling pathways are briefly summarized (
Several stimuli control melanogenesis, including external factors such as UV rays or environmental pollution [11]. Furthermore, a series of endogenous molecules released by melanocytes, keratinocytes, dermal fibroblasts, and immune cells modulate the process paracrinally and autocrinally. These are mainly the α-melanocyte stimulating hormone (α-MSH) and the adrenocorticotropic hormone (ACTH), derived from the cleavage of pro-opiomelanocortin (POMC), from both melanocytes and keratinocytes [11,24]. In addition, stem cell factor (SCF), peptide endothelin 1 (ET-1), hepatocyte growth factor (HGF), keratinocyte growth factor (KGF), basic fibroblast growth factor (bFGF), and inflammatory mediators such as cytokines, prostaglandin E2 (PGE2), and nitric oxide (NO) significantly regulate melanogenesis [25,26]. All these stimuli operate with the activation of several signaling pathways [9,27]. Detailed descriptions of the multiple pathways in melanogenesis have been recently reported by others [6,7,9,11]. Here, the main signaling pathways are briefly summarized ().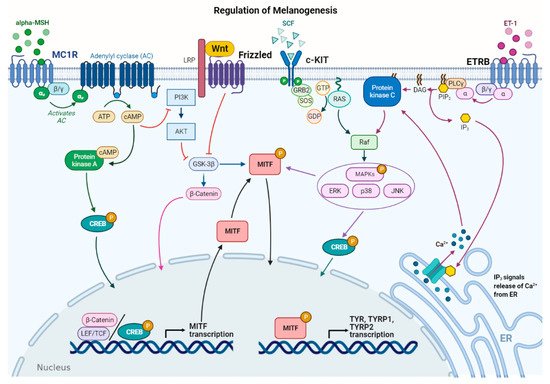
Figure 2.
Main signaling pathways involved in melanogenesis regulation. α-melanocyte stimulating hormone (α-MSH) binds to melanocortin 1 receptor (MC1R), which increases cAMP levels, activating PKA and PI3K/AKT pathway. The former phosphorylates CREB protein, promotingMITF
transcription; the latter interplays with Wnt/β-catenin pathway by phosphorylating GSK-3β, which, in turn, releases β-catenin to promoteMITF
transcription. Stem cell factor (SCF) binds to c-KIT receptor, activating MAPK pathway and phosphorylating CREB. Peptide endothelin 1 (ET-1) binds to its receptor, ETRB, activating PKC and stimulatingMITF
transcription.MITF
is phosphorylated at the post-transcriptional level to promote transcription of the melanogenic enzymes.In melanocytes, signaling pathways regulating melanogenesis operate through membrane receptors with different molecular activities. These receptors include G protein-coupled receptors (GPCRs) such as the melanocortin-1 receptor (MC1R), which is mainly expressed in melanocytes [27], adrenergic receptors, endothelin type B receptor (ETRB), frizzled receptor (FZD), and tyrosine kinase receptors such as tyrosine kinase receptor KIT, bFGF, and HGF receptors [7][28]. Most signaling pathways lead to the regulation of microphthalmia-associated transcription factor (
In melanocytes, signaling pathways regulating melanogenesis operate through membrane receptors with different molecular activities. These receptors include G protein-coupled receptors (GPCRs) such as the melanocortin-1 receptor (MC1R), which is mainly expressed in melanocytes [28], adrenergic receptors, endothelin type B receptor (ETRB), frizzled receptor (FZD), and tyrosine kinase receptors such as tyrosine kinase receptor KIT, bFGF, and HGF receptors [7,29]. Most signaling pathways lead to the regulation of microphthalmia-associated transcription factor (MITF
), a basic helix–loop–helix leucine zipper (bHLH-ZIP) transcription factor.MITF is the dominant transcription factor in melanogenesis as it controls melanocyte development, survival, and proliferation, as well as the steps involved in melanin synthesis [29][30].
is the dominant transcription factor in melanogenesis as it controls melanocyte development, survival, and proliferation, as well as the steps involved in melanin synthesis [30,31].MITF
induces the transcription of the melanogenic genes, includingTYR
,TYRP-1
, andTYRP-2, by binding to the conserved consensus elements of the promoter regions [30]. Moreover,
, by binding to the conserved consensus elements of the promoter regions [31]. Moreover,MITF regulates several other genes involved in melanogenesis, including those required to control melanosome maturation, traffic, and distribution to keratinocytes [31]. Due to the prominent role of
regulates several other genes involved in melanogenesis, including those required to control melanosome maturation, traffic, and distribution to keratinocytes [32]. Due to the prominent role ofMITF
, regulation of its expression and activity represents a key event in melanogenesis. At the transcriptional level,MITF
expression is regulated by several transcriptional factors that bind theMITF promoter, including cyclic adenosine monophosphate (cAMP)-response element binding protein (CREB), paired box family of transcription factor 3 (PAX3), sex determining region Y-box 9 and 10 (Sox9, Sox10), and Wnt/β-catenin pathway effector lymphoid enhancer-binding factor 1 (LEF-1) [7][30].
promoter, including cyclic adenosine monophosphate (cAMP)-response element binding protein (CREB), paired box family of transcription factor 3 (PAX3), sex determining region Y-box 9 and 10 (Sox9, Sox10), and Wnt/β-catenin pathway effector lymphoid enhancer-binding factor 1 (LEF-1) [7,31].Furthermore,MITF activity is regulated at the post-transcriptional level mainly by phosphorylation [32].
activity is regulated at the post-transcriptional level mainly by phosphorylation [33].MITF may be phosphorylated by several kinases such as MAPK, p38, ribosomal S6 kinase (RSK), and GSK3β [7][11][33][34].
may be phosphorylated by several kinases such as MAPK, p38, ribosomal S6 kinase (RSK), and GSK3β [7,11,34,35].MITF phosphorylation can favor the recruitment of transcriptional coactivator CBP/P300 of CREB, thus increasing the transcription of TYR, TYRP-1, and TYRP-2 melanogenic enzymes [30]. The MAPK signaling pathway can be activated by several extracellular factors, which operate through tyrosine kinase receptors including SCF, a relevant signal produced by keratinocytes and fibroblasts, and HGF and bFGF, mainly produced by keratinocytes [35]. Notably, melanogenesis-related signaling pathways may be positively or negatively modulated by several inflammatory factors that finally regulate skin pigmentation. This regulation is very complex as some molecules have a stimulatory effect on melanogenesis, while others show inhibitory effects. In addition, the production of these inflammatory mediators depends on the communication between several epidermal and dermal cells, as well as specific stimuli inducing inflammation [6].
phosphorylation can favor the recruitment of transcriptional coactivator CBP/P300 of CREB, thus increasing the transcription of TYR, TYRP-1, and TYRP-2 melanogenic enzymes [31]. The MAPK signaling pathway can be activated by several extracellular factors, which operate through tyrosine kinase receptors including SCF, a relevant signal produced by keratinocytes and fibroblasts, and HGF and bFGF, mainly produced by keratinocytes [36]. Notably, melanogenesis-related signaling pathways may be positively or negatively modulated by several inflammatory factors that finally regulate skin pigmentation. This regulation is very complex as some molecules have a stimulatory effect on melanogenesis, while others show inhibitory effects. In addition, the production of these inflammatory mediators depends on the communication between several epidermal and dermal cells, as well as specific stimuli inducing inflammation [6].As mentioned above, UV rays represent external stimuli with significant effects on melanogenic signaling pathways, resulting in increased melanin production. Several cellular and molecular processes are involved in the UV–skin reaction. Indeed, UV irradiation inducesMITF expression and consequently melanogenic gene expression [3][30]. Acquired knowledge shows that UV irradiation influences different skin cell types, including melanocytes, keratinocytes, and dermal fibroblasts. UV-induced melanogenesis has been associated with the release of several molecules, including α-MSH, ACTH, SCF, and ET-1, which regulate the signaling pathways described above [24][35][36][37].
expression and consequently melanogenic gene expression [3,31]. Acquired knowledge shows that UV irradiation influences different skin cell types, including melanocytes, keratinocytes, and dermal fibroblasts. UV-induced melanogenesis has been associated with the release of several molecules, including α-MSH, ACTH, SCF, and ET-1, which regulate the signaling pathways described above [25,36,37,38].4. miRNA Activities and Identification
MicroRNAs (miRNAs) represent a class of non-coding RNAs of approximately 21–23 nucleotides in length. Since their discovery in 1993 [38], miRNAs have been largely studied as essential regulators of gene expression. miRNAs are highly conserved among species and are found in different cell types and organisms, including plants, animals, and viruses [39][40]. It is now estimated that miRNAs target approximately 60% of genes in
MicroRNAs (miRNAs) represent a class of non-coding RNAs of approximately 21–23 nucleotides in length. Since their discovery in 1993 [39], miRNAs have been largely studied as essential regulators of gene expression. miRNAs are highly conserved among species and are found in different cell types and organisms, including plants, animals, and viruses [40,41]. It is now estimated that miRNAs target approximately 60% of genes inhumans and other mammals [41]. Due to their role in mRNA expression regulation, miRNAs are involved in all cellular activities, including proliferation, differentiation, migration, apoptosis, and immune responses, both in normal and pathological conditions [42][43][44][45]. Several mechanisms ensure high stability in miRNAs. Since they can be easily detected in almost all bodily fluids, miRNAs are now considered important biological markers and potential therapeutic molecules [46]. Inside cells, miRNAs derive from long precursor transcripts which give rise to mature miRNAs through a multi-step process and are then incorporated into an RNA–protein complex known as the RNA-Induced Silencing Complex (RISC) [47]. miRNA activity mainly induces gene expression reduction by binding to sequences in the 3′ untranslated region (3′-UTR) of target genes. This can lead to mRNA target degradation, or inhibition of translation and reduction in protein levels [44]. However, miRNA activities show higher complexity as each miRNA can target different genes and several miRNAs can regulate the same gene’s expression. Moreover, the strict interplay between long non-coding RNAs (lncRNAs) and miRNAs contributes to gene expression regulation [39][44][48].
and other mammals [42]. Due to their role in mRNA expression regulation, miRNAs are involved in all cellular activities, including proliferation, differentiation, migration, apoptosis, and immune responses, both in normal and pathological conditions [43,44,45,46]. Several mechanisms ensure high stability in miRNAs. Since they can be easily detected in almost all bodily fluids, miRNAs are now considered important biological markers and potential therapeutic molecules [47]. Inside cells, miRNAs derive from long precursor transcripts which give rise to mature miRNAs through a multi-step process and are then incorporated into an RNA–protein complex known as the RNA-Induced Silencing Complex (RISC) [48]. miRNA activity mainly induces gene expression reduction by binding to sequences in the 3′ untranslated region (3′-UTR) of target genes. This can lead to mRNA target degradation, or inhibition of translation and reduction in protein levels [45]. However, miRNA activities show higher complexity as each miRNA can target different genes and several miRNAs can regulate the same gene’s expression. Moreover, the strict interplay between long non-coding RNAs (lncRNAs) and miRNAs contributes to gene expression regulation [40,45,49].In general, from a methodological point of view, investigations into miRNA functions and roles in a selected process include a preliminary phase of miRNA identification using high-throughput methods such as microarray, followed by target gene identification with a bioinformatics approach. Finally, target genes need to be validated by 3′-UTR reporter assays and analysis conducted on induced cellular effects following transfection experiments with miRNA mimics and antagomiRs [49]. miRNAs are involved in skin development and functions both in normal and pathological conditions [50]. Furthermore, increasing evidence shows that selected miRNAs regulate specific events occurring in the skin, such as melanogenesis, as detailed below [51][52].
In general, from a methodological point of view, investigations into miRNA functions and roles in a selected process include a preliminary phase of miRNA identification using high-throughput methods such as microarray, followed by target gene identification with a bioinformatics approach. Finally, target genes need to be validated by 3′-UTR reporter assays and analysis conducted on induced cellular effects following transfection experiments with miRNA mimics and antagomiRs [50]. miRNAs are involved in skin development and functions both in normal and pathological conditions [16]. Furthermore, increasing evidence shows that selected miRNAs regulate specific events occurring in the skin, such as melanogenesis, as detailed below [51,52].4.1. miRNA Regulating MITF
Due to the key role ofMITF
as an essential regulator of the melanogenic process, several studies have investigated miRNAs with potential effects onMITF expression and consequent regulation of mRNA levels in melanogenic enzymes [3][29][30][53] (
expression and consequent regulation of mRNA levels in melanogenic enzymes [3,30,31,53] (). Some preliminary studies concerning melanogenesis regulation by miRNAs have been performed in fiber-producing animals, such asalpaca
, due to the contribution of melanin synthesis to coat color and the specific interest ofalpaca
breeders in animal color coat modulation. Differences in miRNA profiles fromalpaca skins with different colored coats were identified and most differentially expressed miRNAs showed predicted targets involved in pigmentation [54][55]. These results led to further investigation concerning the functional role of selected miRNAs. Data obtained by Zhu Z et al. (2010) demonstrated the functional role of miR-25 in reducing
skins with different colored coats were identified and most differentially expressed miRNAs showed predicted targets involved in pigmentation [54,55]. These results led to further investigation concerning the functional role of selected miRNAs. Data obtained by Zhu Z et al. (2010) demonstrated the functional role of miR-25 in reducingMITF
mRNA and protein,TYR
, andTYRP-1
expression in cultured melanocytes. In addition, an inverse relationship was observed between miR-25 level and coat color. Similarly, inalpaca
melanocytes, miR508-3p also directly targetsMITF
, binding to the 3′-UTR of the gene. miR508-3p overexpression downregulatedMITF
expression, resulting in a decrease in TYR, TYRP-2, and melanin production [56]. Interestingly, in 2012, Dong C. et al. investigated the role of miR-137, another miR-targetingMITF
, in a transgenic mice model [57]. Initially investigating melanoma cells lines, where its overexpression downregulatedMITF
[58], it was found that miR-137 also decreased the expression of the MITF protein and its downstream genes in transgenic mice. Notably, miR-137 had an impact on coat color, demonstrating that modulating a specific miR may significantly regulate melanogenesis, at least in animal models [57].Table 1.
microRNAs regulating MITF in melanogenesis.miRNA Cell Model Target Gene Effect on
MelanogenesisRef. miR-25 Alpaca melanocytes MITF Negative [56] miR508-3p Alpaca melanocytes MITF Negative [56] miR-137 Alpaca melanocytes MITF Negative [58] miR-675 Melanocytes of melasma patients, keratinocytes of melasma MITF Negative [5][59][60][5,59,60] miR-218 [64], has been also investigated in immortalizedhuman
epidermal melanocytes (Pig-1), where it downregulatesMITF
expression and melanin synthesis [63]. Recently, miR-141-3p and miR-200a-3p have been identified asMITF
regulators [52]. In this study, comparing miRNA expression profiles in B16-4A5mouse
melanoma cells which were treated or untreated with α-MSH led to 13 miRNAs being identified as differentially expressed through miRNA array analysis. miR-141-3p, miR-200a-3p, and miR-148a-3p, which targetMITF
, were downregulated in α-MSH-stimulated cells when compared to untreated cells. Furthermore, miR-141-3p and miR-200a-3p overexpression suppressedMITF
expression andTYR
activity in B16-4A5 cells. Notably, the inhibitory effect on melanin synthesis was confirmed in a three-dimensional tissue culture model of thehuman
epidermis (3D-MHE model) [52].4.2. miRNAs Regulating Other Genes in Melanogenesis
Besides the abovementioned studied group of miRNAs, with significant regulatory roles inMITF
, other miRNAs are involved in melanogenesis by regulating the expression of other molecular targets, including melanogenic enzymes, transcription factors, or components of the signaling pathways which regulate melanogenesis ().Several miRNAs, including miR-450b-5p, miR-1208, miR-326, miR-434-5p, miR330-5p, miR-125, miR-145, and miR-203, have been predicted as targetingTYR
and most also target other genes, including[65,66].Table 2.
Other microRNAs involved in melanogenesis.miRNA Cell Model Target Gene Effect on
MelanogenesisRef. miR-434-5p Mouse skin, human skin cell cultures TYR Negative [65] miR-330-5p Melanoma cells, normal human melanocytes TYR Negative [56][67][56,67] miR-203 Keratinocytes exposed to UV Kinesin Superfamily Protein 5b Positive [67] miR-3196 Keratinocytes exposed to UV Unknown target gene Positive [67] Melan-a murine melanocytes, human skin OTC MITF Negative [61] miR-21a-5p Human melanocytes SOX5 Positive [68] miR-183 B16 melanoma cells MITF Negative [62] miR-340 Human epidermal melanocytes (Pig-I) MITF Negative [63][64][63,64] miR-200a-3p B16-4A5 melanoma cells MITF Negative [52] miR-148a-3p B16-4A5 melanoma cells MITF Negative [52] miR-141-3p B16-4A5 melanoma cells MITF Negative [52] Other studies have identified miRNAs regulating skin pigmentation inhuman
melanocytes. Particular interest has been shown in miR-675, another miR which targetsMITF and was to be found expressed at low levels in the hyperpigmented skin of melasma patients. In melanocytes or keratinocytes derived from melasma patients, miR-675 upregulation decreased TYRP-1 and TYRP-2 expression, whereas its knockdown increased their expression. Interestingly, miR-675 was also identified in exosomes released from keratinocytes into the extracellular environment [5][59][60]. Another miR involved in melanogenesis by binding the 3′-UTR in
and was to be found expressed at low levels in the hyperpigmented skin of melasma patients. In melanocytes or keratinocytes derived from melasma patients, miR-675 upregulation decreased TYRP-1 and TYRP-2 expression, whereas its knockdown increased their expression. Interestingly, miR-675 was also identified in exosomes released from keratinocytes into the extracellular environment [5,59,60]. Another miR involved in melanogenesis by binding the 3′-UTR inMITF
is miR-218, which downregulated TYR, TYRP-1, and DCT mRNA and protein levels, reducing melanin content in immortalized melan-a murine melanocytes. In agreement with these data, miR-218 also suppressed melanogenesis inhuman
pigmented skin organotypic culture (OTC) throughMITF
repression. Anti-miR-218 was also found to promote melanogenesis inhuman
primary melanocytes [61].Recently, other miRNAs, including miRNA-183 cluster, miR-340, miR-141-3p, and miR-200a-3p, have been investigated for their role in regulatingMITF
expression and melanogenesis [62]. Results of this research show that the miRNA-183 cluster targets the 3′-UTR ofMITF
in B16mouse
melanoma cells. miRNA-183 cluster overexpression decreasedMITF
,TYR
,TYRP-1
, andDCT
expression and melanin production. Conversely, knockdown of the miRNA-183 cluster increasedMITF
,TYR
,TYRP-1
, and DCT expression and, consequently, melanin levels. The miR-183 cluster was also involved in regulating the MEK/ERK signaling pathway implicated in cell proliferation and migration, by modulating mitogen-activated protein kinase 1 (MEK1), extracellular regulated protein kinases1/2 (ERK1/2), and CREB expression [62]. miR-340, which was first identified in melanoma cell lines for its ability to bind specifically to the 3′-UTR ofMITF
miR-145 Murine melan-a melanocytes Myo5a Negative [ 56 ] miR-380-3p Alpaca melanocytes SOX6 Negative [56] miR-200c Normal human epidermal keratinocytes (NHEK) SOX1 Positive [69] miR-27a-3p Alpaca and Mouse melanocytes Wnt3a Negative [54][70][54,70] miR-379 Alpaca melanocytes IGF1R Negative [71] miR-143-5p Human melanocytes Myo5a Negative [72] miR-143-5p Alpaca melanocytes TAK1 Negative [73] miR-125b WM266-4 human melanoma cells, MNT1 human melanoma cells SH3BP4 Negative [60] Wu et al. used a miR-434-5p homologue to targetTYR
in cells cultured in vitro as well as in an animal model and showed efficient melanin synthesis reduction. Similarly, miR-330-5p downregulatedTYR
in melanoma cells and normal melanocytes, inducing depigmentation without affecting cell proliferation [66]. This miR has been also identified in exosomes derived from keratinocytes [56]. In this study, exosomes carrying miR-330-5p caused a decrease in melanin production andTYR
expression in melanocytes. Similarly, miR-330-5p overexpression in melanocytes confirmed its inhibitory activity on melanogenesis. Several other miRNAs, such as miR-203, which targetsKinesin Superfamily Protein 5b
, involved in melanosome transfer, and miR-3196, with unknown target genes, have been identified inhuman
-derived exosomes from keratinocytes and have shown an ability to increase melanin content in melanocytes [67]. Indeed, exosomes released by Black keratinocytes, as well as Caucasian UV-irradiated keratinocytes, were able to induce increased TYR activity and melanogenic gene expression in melanocytes, at least partly as a result of miR content.As reported above, several miRNAs which directly targetMITF
are involved in melanogenesis. Conversely,MITF
expression may be subject to more complex regulation. For example, it has been shown that miR-21a-5p overexpression downregulated theSOX5
target, as well as β-catenin and CDK2 protein expression, in normalhuman
melanocytes. In agreement with
