You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Petros Christopoulos.
The principles and current clinical landscape of multispecific antibodies against cancer.
- bispecific antibodies
- multispecific antibodies
- monoclonal antibodies
- therapeutic antibodies
1. Introduction
The tortuous, 2.5 billion-years-long path from immunoglobulin (Ig)-like domains of archaeal flagellins to the antibodies (Ab) of jawed vertebrates is one of the most intriguing discoveries in evolutionary biology [1,2,3][1][2][3]. Equally impressive are the accomplishments of modern genetic engineering, whose further variation of basic Ig building blocks could produce over 40 different molecular formats of therapeutic antibodies during the last two decades [4]. As times change, priorities shift, and pathogen defense has today largely been succeeded by a much more challenging task: the fight against cancer [5].
The concept that antibodies could be used as “magic bullets” against human maladies dates back to their discovery in the late 19th century [6] and the gradual recognition that they can bind a virtually unlimited number of antigens with a high specificity and affinity [7]. However, it was not until discovery of the “hybridoma” technology in 1975 [8], complemented by various humanization techniques a few years later [9], that scientists managed to harness this power: monoclonal antibodies could now be produced in large quantities after injecting a mouse (later, rat or other mammal) with the desired antigen, and fusing the respective splenocytes with suitable myeloma cell lines [10]. In the next step, two hybridomas were fused (“hybrid hybridoma”, aka “quadroma”) to produce bispecific antibodies without the protein denaturation steps necessary for chemical cross-linking, which could potentially adversely affect binding properties [11,12][11][12].
Improved function is the main incentive behind development of multispecific constructs. For every antibody, the “classic” mode of action falls into two broad categories: (i) “disruptive” with respect to the epitope-bearing target molecule, i.e., blocking or activating signals, neutralizing antigens, or causing internalization and degradation of surface receptors and (ii) “recruiting”, i.e., activating immune cells and/or other effector molecules, like the complement [13]. OKT3 (aka “muromonab-CD3”), for example, the first monoclonal antibody to ever achieve regulatory approval in 1986, is a typical product of the first category, used to suppress T-cell function in patients with glucocorticoid-resistant rejection of allogeneic renal, heart and liver transplants [14]. Rituximab, on the other hand, a CD20-specific monoclonal antibody still widely used since its approval in 1997, kills B- cells by combining signaling-induced death with cellular and complement-mediated cytotoxicity [15]. Compared to monospecific monoclonal antibodies, multispecific constructs potentiate antibody-mediated effects, for example, they can potentially “disrupt” multiple instead of one tumor-associated antigens (TAA) owing to more antigen-binding regions, and/or they can “recruit” and activate immune cells even stronger, since they use dedicated antigen-binding sites for this. The functional augmentation facilitated by multispecificity is clinically relevant: it translates into improved response rates, for example, approximately 50% with the newer anti-CD20/CD3 bispecific antibodies as monotherapy in B-cell non-Hodgkin’s lymphomas (B-NHL) which do not respond to rituximab any more [16], can delay development of resistance, and simplifies drug development compared to the more complicated, expensive and time-consuming procedures necessary for launching of multiple monospecific products instead [4].
Wide adoption of genetic engineering facilitates today’s exploitation of the huge potential inherent in multispecific antibodies: suitable polypeptide chains are designed in silico and expressed in various host systems, most frequently CHO cells and E. coli, followed by purification and assembly of the various components in vitro [17,18,19][17][18][19]. Appropriate antigen-binding properties, high yield, high thermal and chemical stability, good solubility, and low viscosity have key importance for large-scale production and clinical applicability [20,21][20][21]. The biochemical basis of these characteristics and our ability to manipulate them lie rooted in the modular antibody structure (Figure 1a).
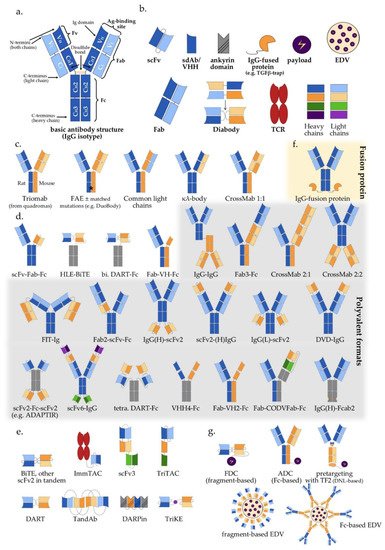
Figure 1. Multispecific antibody formats in clinical trials: (a) Basic IgG structure; (b) Main antibody components (detailed in Section 2 of the main text); (c) IgG-like “two-halves” bispecific formats (following the order of Section 3.1 and Table 1); (d) IgG-modified (appended and/or substituted, Fc-based) bispecific and multispecific antibodies (Table 1); (e) Fragment-based (Fc-free) bispecific and multispecific antibodies (Table 1); (f) IgG fusion protein; (g) Bi-/multispecific constructs for payload delivery (Table 1). For the abbreviations, please see the respective section in the main text.
Table 1. Multispecific antibody formats in clinical trials by structure, mode of action and start year of first study.
| Class | Specificity/ Valence |
Specificity/ Valence |
Action (C/R) | Format | No. of Clinical Trials | Comment | First Clinical Trial |
|---|
| Fc-based | IgG-like (Section 3.1) (Figure 1c) | (Figure 1c) |
2/2 | R | TrioMab | 9 | “two-halves” formats |
2004 | ||
| 2/2 | C/R | FAE | 36 | 2010 | ||||||
| 2/2 | C/R | common light chain | 19 | 2014 | ||||||
| 2/2 | C | κλ-body | 2 | 2019 | ||||||
| 2/2 | C/R | CrossMab 1:1 | 12 | 2012 | ||||||
| IgG-modified (Section 3.2 & Section 4) (Figure 1d) | (Figure 1d) |
2/2 | C/R | scFv-Fab-Fc | 18 | scFv-monosubstituted | 2016 | |||
| 2/2 | R | HLE-BiTE | 11 | scFv-bisubstituted | 2015 | |||||
| 2/2 | R | DART-Fc | 2 | Db-bisubstituted | 2014 | |||||
| 2/2 | R | Fab-VH-Fc ** | 3 | V | H | -monosubstituted | 2021 | |||
| 2/4 | R | IgG-IgG | 15 | IgG-IgG | 2004 | |||||
| 2/3 | R | Fab3-Fc | 1 | Fab-appended | 2018 | |||||
| 2/3 | R | CrossMab 2:1 | 10 | 2014 | ||||||
| 2/4 | C | CrossMab 2:2 | 1 | 2015 | ||||||
| 2/4 | C/R | FIT-Ig (Fabs-in-Tandem) | 3 | 2018 | ||||||
| 2/3 | R | Fab2-scFv-Fc | 2 | scFv-appended | 2020 | |||||
| 2/4 | C/R | IgG(H)-scFv2 | 19 | 2017 | ||||||
| 2/4 | C | scFv2-(H)IgG | 1 | 2014 | ||||||
| 2/4 | R | IgG(L)-scFv2 | 2 | 2019 | ||||||
| 2/4 | C | DVD-IgG | 1 | V-appended | 2013 | |||||
| 2/4 | R | scFv2-Fc-scFv2 | 2 | scFv-multisubstituted | 2015 | |||||
| 4/8 | R | scFv6-IgG | 2 | 2020 | ||||||
| 2/4 | C | DART-Fc | 3 | Db-multisubstituted | 2017 | |||||
| 2/4 | C | VHH4-Fc | 5 | V-multi substituted | 2019 | |||||
| 2/3 | C/R | Fab-VH2-Fc ** | 4 | 2019 | ||||||
| 3/3 | R | Fab-CODVFab-Fc | 1 | 2020 | ||||||
| 2/3-4 * | C | IgG-fusion proteins | 55 | Fusion moiety | 2015 | |||||
| 2/4 | C | IgG(H)-Fcab2 | 3 | Fc-modified | 2018 | |||||
| Fc-free | Fv-based $ | ( | $Section 3.3 & Section 4) ($ Figure 1e) | Figure 1e) | 2/2 | R | BiTE | 20 | scFv-based | 2008 |
| 2/2 | C/R | other scFv2 in tandem | 6 | 2005 | ||||||
| 2/2 * | R | ImmTAC | 5 | 2015 | ||||||
| 3/3 * | R | TriKE | 1 | 2020 | ||||||
| 3/3 | C | scFv3 | 1 | 2020 | ||||||
| 2/2 | R | DART | 6 | Db-based | 2014 | |||||
| 2/4 | R | TandAb | 6 | 2010 | ||||||
| 3/3 | R | TriTAC | 4 | 2018 | ||||||
| ¾ * | C | DARPin | 5 | Ankyrin-based | 2014 | |||||
| Fab-based | $ | 2/2 | C | Fab2 | 2 | Fab-based | 1997 | |||
| with payload (Section 5) (Figure 1f) | (Figure 1f) |
2/2 # | C | Fc-based ADC/EDV | 4 | IgG-based | 2014 | |||
| 2/2 # | C | Fc-free FDC/EDV | 3 | Fragment-based | 2013 | |||||
| 2/2-3 (#) | C | ±pretargeting (±imaging) | 4 | 2004 | ||||||
Formats are ordered as in Figure Figure 11 and the corresponding sections of the main text; C: classical mode of action; R: immune-cell redirecting; for the explanation of other abbreviations, please see the main text; * one binding site does not rely on typical antigen-antibody interaction; ** human VH or VHH; # with payload; $ Figure 1e.
2. Antibody Structure and Approaches to Multispecificity
The typical structure of human antibodies is represented by the IgG isotype, which is the most prevalent class [22]. X-ray crystallographic and electron-microscopic studies have revealed this to be a heterotetrametric, roughly Y-shaped protein with axial symmetry, which consists of two identical heavy (“H”, approximately 50 kDa each), and two identical “light” (“L”, approximately 25 kDa each) polypeptide chains, linked together by disulfide bonds (Figure 1a) [23,24,25][23][24][25]. Basic building block for both chain types is the “Ig domain”, aka “Ig fold”, which is a sandwich-like structure formed by two sheets of 7–9 antiparallel β-strands arranged with a Greek-key topology [26]. This basic motif appears to be conserved throughout the evolution of life, presumably because its efficient and compact folding provides a suitable substrate for numerous essential recognition, binding and adhesion processes carried out by members of the large Ig protein superfamily [1]. Each human antibody heavy chain consists of four domains, three constant ones (termed CH1, CH2 and CH3) and one variable (VH), while the light chains consist of one constant (CL) and one variable domain (VL) each. Limited digestion with the cysteine protease papain splits the IgG antibody in three equal-sized portions, namely two antigen-binding fragments (Fab), each consisting of one light chain bound with the VH and CH1 domains of its partner heavy chain, and one crystallizable fragment (Fc), which contains the remaining constant domains of the heavy chain (CH2 and CH3, Figure 1a) [27]. Within the Fab region, the side-by-side arrangement of the VL and VH domains brings discrete amino acid loops between their β-sheets (“complementarity-determining regions”, CDR) together to form the antigen-binding site at the outer tip (Figure 1a). Direct linking of VL and VH by a peptide chain creates the single-chain variable fragment (scFv), an artificial construct that can also effectively bind antigens (Figure 1b) [28]. Interestingly, the antigen specificity of most antibodies shows a predominant dependence on the CDRs contributed by mainly the VH rather than the VL domain, which takes an extreme form in the heavy chain-only antibodies naturally produced by camelids and sharks [29], and is exploited by “single-domain antibodies” (sdAb), also referred to as nanobodies, consisting of a single VHH (variable heavy chain homodimer) thereof (Figure 1b) [30].
Due to its axial symmetry, the IgG can bind two identical epitopes with two binding sites (“paratopes”), one on each Fab arm, i.e., it is “monospecific”, but “bivalent” [31]. Generally, the valency of an antibody refers to the total number of epitopes that it can bind, and its specificity to the number of different structures among them. Naturally occurring antibodies are generally monospecific, which allows them to cross-link large numbers of antigen molecules and thus amplify the immune response against them [22]. Nonetheless, for IgG4 and possibly also other Ig subclasses, reducing conditions in the blood or cell surfaces can break up disulfide bonds and facilitate Fab-arm exchange (FAE) which gives chimeric bispecific antibodies with anti-inflammatory activity (Figure 1c) [32,33][32][33]. Generation of the first artificial bispecific antibodies in 1961 mimicked this naturally occurring FAE: by reducing and reoxidizing the F(ab’)2 fragments derived by peptic digestion of two different antibodies, their univalent Fab were recombined into new F(ab’)2 molecules that could precipitate a mixture of their cognate antigens, but neither of them in pure form [34,35][34][35]. After 1975, “quadromas” became a relatively easy method to generate full IgG-like bispecific molecules (Figure 1c), but the low yield of the desired antibody (12.5% with random pairing of heavy and light chains), together with the difficulty to isolate it from the closely related mispaired contaminants, remained a significant problem [36]. Over the following decades, many different methods within the constraints of the classical “IgG-like” format were devised to overcome or circumvent the “mispairing” problem of heavy and light chains produced by quadromas or genetic engineering (Figure 1c): rat/mouse quadromas, which took advantage of the species-restricted heavy/light chain pairing and the differential affinity of protein A for mouse and rat heavy chains (“TrioMabs”) [37,38][37][38]; various “knobs-into-holes” (KiH) or other techniques of inducing complementary mutations in the sequences of heavy and/or light chains in order to force the desired heterodimerization [39,40,41,42][39][40][41][42] or facilitate controlled FAE (“Duobodies”) [43]; “common-light-chain” antibodies, in which the two distinct paratopes on each Fab arm utilize the same light chain paired with a different heavy chain in order to bind its target antigen [44]; “κ-λ” bodies, which utilize the same heavy, but different light chains for the two paratopes, in order to obviate the need for artificial mutations or linkers that may result in poor stability and increased potential immunogenicity [45]; “CrossMabs”, with swapping of either the variable or the constant domains between light and heavy chains to create two asymmetric Fab arms that force the desired light chain pairing, while preserving the binding properties of the respective paratope [46,47][46][47]; electrostatic steering effects [48,49][48][49]; IgG/A chimeras, aka “strand exchange engineered domain bodies” (“SEEDBodies”) [50]; the proprietary “Azymetric” heterodimeric Fc [51]; “dual action Fab” (DAF, aka “two-in-one” antibodies), which use the same heavy and light chains to recognize two unrelated antigens via differential use of their two paratopes [52,53][52][53]; “DutaMabs” or “DutaFabs”, in which each Fab arm contains two different paratopes, each utilizing only 3 out of the 6 available CDRs [54].
“IgG-modified” formats (Figure 1d) provide additional solutions to this problem, e.g., in “dual-variable-domain” (DVD) antibodies, each chain contains two variable domains, so that bispecificity is ensured irrespective of light chain pairing [55]. Moreover, using genetic engineering, the antigen-binding moieties Fab, Fv and VHH can be combined freely with each other, or linked to IgGs, resulting in huge structural variability (Figure 1d,e and Table 1) [4,56][4][56]. In addition, monospecific and bispecific formats can be combined in order to increase specificity, valency, or both; for example a highly active tetravalent and tetraspecific “four-in-one” antibody against EGFR, HER2, HER3 and VEGF was generated by combining the DVD, CrossMab and KiH technologies [57]. It should also be noted that the functionality of multispecifics can further be extended through fusion with other non-Ig proteins; for example, the T-cell receptor (TCR) [58], the TGFβ receptor [59], an IL-15 moiety in case of Trispecific Killer Cell Engagers (TriKEs) [60], various payloads [61], and by conjugation with Engeneic Delivery Vehicles (EDV), i.e., bacterially-derived nanocells coated with bispecific antibodies for the targeted delivery of cytotoxics, siRNA and other cargo to the tumor cells (Figure 1f) [62,63][62][63].
Among the numerous possible variations, the single most important structural characteristic of multispecific antibodies is whether they contain an Fc region or not. Fc-based constructs are generally larger, have longer half-lives (typically a few weeks) due to recycling by the neonatal Fc receptor (FcRn) and glomerular preservation [64[64][65],65], show improved solubility and stability, trigger cytotoxic [66] and T-cell priming effects [67], and can be purified using established affinity chromatography workflows [68]. In contrast, Fc-free, antibody fragment-based constructs are usually rapidly cleared from the blood (within minutes), which necessitates either administration by continuous infusion, or fusion with carrier moieties, such as human serum albumin (HSA) or polyethylene glycol (PEG), in order to extend their half-lives [69,70,71][69][70][71]. At the same time, specific advantages of smaller multispecifics can be better diffusion into the tumor tissue, higher potency due to closer proximity of interactions in the two paratopes, ease of large-scale production in microbial systems, and less immune-related adverse effects due to lack of Fc [72,73,74][72][73][74].
In terms of functionality, the basic distinction is between the “classic” and the “recruiting” or “redirecting” mode of antibody action, in which at least one binding site engages invariable immune cell receptors, for example, CD3 on T, or CD16 on NK cells. The combined structural (“Fc-based” vs. “Fc-free”) and functional (“classic” vs. “recruiting”) characterization is a useful framework to contextualize multispecific constructs currently used in clinical trials (Table 1). A detailed description of all constructs and corresponding studies can be found in the full text and supplements of the original IJMS publication.
References
- Oreste, U.; Ametrano, A.; Coscia, M.R. On Origin and Evolution of the Antibody Molecule. Biology 2021, 10, 140.
- Braun, T.; Vos, M.R.; Kalisman, N.; Sherman, N.E.; Rachel, R.; Wirth, R.; Schröder, G.F.; Egelman, E.H. Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc. Natl. Acad. Sci. USA 2016, 113, 10352–10357.
- Tilson, M.D.; Rzhetsky, A. A Novel Hypothesis Regarding the Evolutionary Origins of the Immunoglobulin Fold. Curr. Med. Res. Opin. 2000, 16, 88–93.
- Spiess, C.; Zhai, Q.; Carter, P.J. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol. Immunol. 2015, 67, 95–106.
- Souriau, C.; Hudson, P.J. Recombinant antibodies for cancer diagnosis and therapy. Expert Opin. Biol. Ther. 2003, 3, 305–318.
- Scott, A.M.; Allison, J.P.; Wolchok, J.D. Monoclonal antibodies in cancer therapy. Cancer Immun. 2012, 12, 14.
- Rabia, L.A.; Desai, A.A.; Jhajj, H.S.; Tessier, P.M. Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem. Engin. J. 2018, 137, 365–374.
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497.
- Safdari, Y.; Farajnia, S.; Asgharzadeh, M.; Khalili, M. Antibody humanization methods—A review and update. Biotechnol. Genet. Eng. Rev. 2013, 29, 175–186.
- Parray, H.A.; Shukla, S.; Samal, S.; Shrivastava, T.; Ahmed, S.; Sharma, C.; Kumar, R. Hybridoma technology a versatile method for isolation of monoclonal antibodies, its applicability across species, limitations, advancement and future perspectives. Int. Immunopharmacol. 2020, 85, 106639.
- Margulies, D.H.; Michael Kuehl, W.; Scharff, M.D. Somatic cell hybridization of mouse myeloma cells. Cell 1976, 8, 405–415.
- Milstein, C.; Cuello, A.C. Hybrid hybridomas and their use in immunohistochemistry. Nature 1983, 305, 537–540.
- Casali, P.; Schettino, E.W. Structure and function of natural antibodies. Curr. Top Microbiol. Immunol. 1996, 210, 167–179.
- Norman, D.J. Mechanisms of action and overview of OKT3. Ther. Drug Monit. 1995, 17, 615–620.
- Weiner, G.J. Rituximab: Mechanism of action. Semin. Hematol. 2010, 47, 115–123.
- Lussana, F.; Gritti, G.; Rambaldi, A. Immunotherapy of Acute Lymphoblastic Leukemia and Lymphoma with T Cell–Redirected Bispecific Antibodies. JCO 2021, 39, 444–455.
- Spiess, C.; Merchant, M.; Huang, A.; Zheng, Z.; Yang, N.-Y.; Peng, J.; Ellerman, D.; Shatz, W.; Reilly, D.; Yansura, D.G.; et al. Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat. Biotechnol. 2013, 31, 753–758.
- Wang, Q.; Chen, Y.; Park, J.; Liu, X.; Hu, Y.; Wang, T.; McFarland, K.; Betenbaugh, M.J. Design and Production of Bispecific Antibodies. Antibodies 2019, 8, 43.
- Chon, J.H.; Zarbis-Papastoitsis, G. Advances in the production and downstream processing of antibodies. Nat. Biotechnol. 2011, 28, 458–463.
- Demarest, S.J.; Glaser, S.M. Antibody therapeutics, antibody engineering, and the merits of protein stability. Curr. Opin. Drug Discov. Dev. 2008, 11, 675–687.
- Kelley, B. Industrialization of mAb production technology: The bioprocessing industry at a crossroads. mAbs 2009, 1, 443–452.
- Schroeder, H.W.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52.
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 2014, 5, 520.
- Silverton, E.W.; Navia, M.A.; Davies, D.R. Three-dimensional structure of an intact human immunoglobulin. Proc. Natl. Acad. Sci. USA 1977, 74, 5140–5144.
- Fleischman, J.B.; Porter, R.R.; Press, E.M. The arrangement of the peptide chains in gamma-globulin. Biochem. J. 1963, 88, 220–228.
- Bork, P.; Holm, L.; Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994, 242, 309–320.
- Porter, R.R. The hydrolysis of rabbit y-globulin and antibodies with crystalline papain. Biochem. J. 1959, 73, 119–126.
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250.
- Bever, C.S.; Dong, J.-X.; Vasylieva, N.; Barnych, B.; Cui, Y.; Xu, Z.-L.; Hammock, B.D.; Gee, S.J. VHH antibodies: Emerging reagents for the analysis of environmental chemicals. Anal. Bioanal. Chem. 2016, 408, 5985–6002.
- Harmsen, M.M.; de Haard, H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007, 77, 13–22.
- Heidelberger, M. The Molecular Composition of Specific Immune Precipitates from Rabbit Sera. J. Am. Chem. Soc. 1938, 60, 242.
- Van der Neut Kolfschoten, M.; Schuurman, J.; Losen, M.; Bleeker, W.K.; Martínez-Martínez, P.; Vermeulen, E.; den Bleker, T.H.; Wiegman, L.; Vink, T.; Aarden, L.A.; et al. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science 2007, 317, 1554–1557.
- Sedykh, S.E.; Lekchnov, E.A.; Prince, V.V.; Buneva, V.N.; Nevinsky, G.A. Half molecular exchange of IgGs in the blood of healthy humans: Chimeric lambda-kappa-immunoglobulins containing HL fragments of antibodies of different subclasses (IgG1-IgG4). Mol. Biosyst. 2016, 12, 3186–3195.
- Nisonoff, A.; Rivers, M.M. Recombination of a mixture of univalent antibody fragments of different specificity. Arch. Biochem. Biophys. 1961, 93, 460–462.
- Nisonoff, A.; Wissler, F.C.; Lipman, L.N. Properties of the major component of a peptic digest of rabbit antibody. Science 1960, 132, 1770–1771.
- Suresh, M.R.; Cuello, A.C.; Milstein, C. Advantages of bispecific hybridomas in one-step immunocytochemistry and immunoassays. Proc. Natl. Acad. Sci. USA 1986, 83, 7989–7993.
- Lindhofer, H.; Mocikat, R.; Steipe, B.; Thierfelder, S. Preferential species-restricted heavy/light chain pairing in rat/mouse quadromas. Implications for a single-step purification of bispecific antibodies. J. Immunol. 1995, 155, 219–225.
- Ollier, R.; Wassmann, P.; Monney, T.; Ries Fecourt, C.; Gn, S.; Ca, V.; Ayoub, D.; Stutz, C.; Gudi, G.S.; Blein, S. Single-step Protein A and Protein G avidity purification methods to support bispecific antibody discovery and development. mAbs 2019, 11, 1464–1478.
- Ridgway, J.B.; Presta, L.G.; Carter, P. ’Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996, 9, 617–621.
- Atwell, S.; Ridgway, J.B.; Wells, J.A.; Carter, P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J. Mol. Biol. 1997, 270, 26–35.
- Kuglstatter, A.; Stihle, M.; Neumann, C.; Müller, C.; Schaefer, W.; Klein, C.; Benz, J. Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products. Protein Eng. Des. Sel. 2017, 30, 649–656.
- Moore, G.L.; Bautista, C.; Pong, E.; Nguyen, D.-H.T.; Jacinto, J.; Eivazi, A.; Muchhal, U.S.; Karki, S.; Chu, S.Y.; Lazar, G.A. A novel bispecific antibody format enables simultaneous bivalent and monovalent co-engagement of distinct target antigens. mAbs 2011, 3, 546–557.
- Labrijn, A.F.; Meesters, J.I.; de Goeij, B.E.C.G.; van den Bremer, E.T.J.; Neijssen, J.; van Kampen, M.D.; Strumane, K.; Verploegen, S.; Kundu, A.; Gramer, M.J.; et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc. Natl. Acad. Sci. USA 2013, 110, 5145–5150.
- Merchant, A.M.; Zhu, Z.; Yuan, J.Q.; Goddard, A.; Adams, C.W.; Presta, L.G.; Carter, P. An efficient route to human bispecific IgG. Nat. Biotechnol. 1998, 16, 677–681.
- Fischer, N.; Elson, G.; Magistrelli, G.; Dheilly, E.; Fouque, N.; Laurendon, A.; Gueneau, F.; Ravn, U.; Depoisier, J.-F.; Moine, V.; et al. Exploiting light chains for the scalable generation and platform purification of native human bispecific IgG. Nat. Commun. 2015, 6, 6113.
- Klein, C.; Schaefer, W.; Regula, J.T.; Dumontet, C.; Brinkmann, U.; Bacac, M.; Umaña, P. Engineering therapeutic bispecific antibodies using CrossMab technology. Methods 2019, 154, 21–31.
- Fenn, S.; Schiller, C.B.; Griese, J.J.; Duerr, H.; Imhof-Jung, S.; Gassner, C.; Moelleken, J.; Regula, J.T.; Schaefer, W.; Thomas, M.; et al. Crystal structure of an anti-Ang2 CrossFab demonstrates complete structural and functional integrity of the variable domain. PLoS ONE 2013, 8, e61953.
- Gunasekaran, K.; Pentony, M.; Shen, M.; Garrett, L.; Forte, C.; Woodward, A.; Ng, S.B.; Born, T.; Retter, M.; Manchulenko, K.; et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: Applications to bispecific molecules and monovalent IgG. J. Biol. Chem. 2010, 285, 19637–19646.
- Strop, P.; Ho, W.-H.; Boustany, L.M.; Abdiche, Y.N.; Lindquist, K.C.; Farias, S.E.; Rickert, M.; Appah, C.T.; Pascua, E.; Radcliffe, T.; et al. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J. Mol. Biol. 2012, 420, 204–219.
- Davis, J.H.; Aperlo, C.; Li, Y.; Kurosawa, E.; Lan, Y.; Lo, K.-M.; Huston, J.S. SEEDbodies: Fusion proteins based on strand-exchange engineered domain (SEED) CH3 heterodimers in an Fc analogue platform for asymmetric binders or immunofusions and bispecific antibodies. Protein Eng. Des. Sel. 2010, 23, 195–202.
- Von Kreudenstein, T.S.; Escobar-Carbrera, E.; Lario, P.I.; D’Angelo, I.; Brault, K.; Kelly, J.; Durocher, Y.; Baardsnes, J.; Woods, R.J.; Xie, M.H.; et al. Improving biophysical properties of a bispecific antibody scaffold to aid developability: Quality by molecular design. mAbs 2013, 5, 646–654.
- Bostrom, J.; Yu, S.-F.; Kan, D.; Appleton, B.A.; Lee, C.V.; Billeci, K.; Man, W.; Peale, F.; Ross, S.; Wiesmann, C.; et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science 2009, 323, 1610–1614.
- Schaefer, G.; Haber, L.; Crocker, L.M.; Shia, S.; Shao, L.; Dowbenko, D.; Totpal, K.; Wong, A.; Lee, C.V.; Stawicki, S.; et al. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell 2011, 20, 472–486.
- Beckmann, R. Dual Targeting. Patent Application No. 14/122,758, 24 July 2014.
- Wu, C.; Ying, H.; Grinnell, C.; Bryant, S.; Miller, R.; Clabbers, A.; Bose, S.; McCarthy, D.; Zhu, R.-R.; Santora, L.; et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Biotechnol. 2007, 25, 1290–1297.
- Labrijn, A.F.; Janmaat, M.L.; Reichert, J.M.; Parren, P.W.H.I. Bispecific antibodies: A mechanistic review of the pipeline. Nat. Rev. Drug Discov. 2019, 18, 585–608.
- Hu, S.; Fu, W.; Xu, W.; Yang, Y.; Cruz, M.; Berezov, S.D.; Jorissen, D.; Takeda, H.; Zhu, W. Four-in-one antibodies have superior cancer inhibitory activity against EGFR, HER2, HER3, and VEGF through disruption of HER/MET crosstalk. Cancer Res. 2015, 75, 159–170.
- Liddy, N.; Bossi, G.; Adams, K.J.; Lissina, A.; Mahon, T.M.; Hassan, N.J.; Gavarret, J.; Bianchi, F.C.; Pumphrey, N.J.; Ladell, K.; et al. Monoclonal TCR-redirected tumor cell killing. Nat. Med. 2012, 18, 980–987.
- Ravi, R.; Noonan, K.A.; Pham, V.; Bedi, R.; Zhavoronkov, A.; Ozerov, I.V.; Makarev, E.; Artemov, A.V.; Wysocki, P.T.; Mehra, R.; et al. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat. Commun. 2018, 9, 741.
- Felices, M.; Lenvik, T.R.; Davis, Z.B.; Miller, J.S.; Vallera, D.A. Generation of BiKEs and TriKEs to Improve NK Cell-Mediated Targeting of Tumor Cells. Methods Mol. Biol. 2016, 1441, 333–346.
- De Goeij, B.E.C.G.; Vink, T.; Ten Napel, H.; Breij, E.C.W.; Satijn, D.; Wubbolts, R.; Miao, D.; Parren, P.W.H.I. Efficient Payload Delivery by a Bispecific Antibody-Drug Conjugate Targeting HER2 and CD63. Mol. Cancer Ther. 2016, 15, 2688–2697.
- MacDiarmid, J.A.; Mugridge, N.B.; Weiss, J.C.; Phillips, L.; Burn, A.L.; Paulin, R.P.; Haasdyk, J.E.; Dickson, K.-A.; Brahmbhatt, V.N.; Pattison, S.T.; et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell 2007, 11, 431–445.
- EDV Technology—EnGeneIC. Available online: (accessed on 21 March 2021).
- Pyzik, M.; Sand, K.M.K.; Hubbard, J.J.; Andersen, J.T.; Sandlie, I.; Blumberg, R.S. The Neonatal Fc Receptor (FcRn): A Misnomer? Front. Immunol. 2019, 10, 1540.
- Kontermann, R.E. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs 2009, 23, 93–109.
- Kellner, C.; Derer, S.; Valerius, T.; Peipp, M. Boosting ADCC and CDC activity by Fc engineering and evaluation of antibody effector functions. Methods 2014, 65, 105–113.
- Ruf, P.; Lindhofer, H. Induction of a long-lasting antitumor immunity by a trifunctional bispecific antibody. Blood 2001, 98, 2526–2534.
- Arora, S.; Saxena, V.; Ayyar, B.V. Affinity chromatography: A versatile technique for antibody purification. Methods 2017, 116, 84–94.
- Kontermann, R.E. Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 2011, 22, 868–876.
- Van Witteloostuijn, S.B.; Pedersen, S.L.; Jensen, K.J. Half-Life Extension of Biopharmaceuticals using Chemical Methods: Alternatives to PEGylation. ChemMedChem 2016, 11, 2474–2495.
- Lorenczewski, G.; Friedrich, M.; Kischel, R.; Dahlhoff, C.; Anlahr, J.; Balazs, M.; Rock, D.; Boyle, M.C.; Goldstein, R.; Coxon, A.; et al. Generation of a Half-Life Extended Anti-CD19 BiTE® Antibody Construct Compatible with Once-Weekly Dosing for Treatment of CD19-Positive Malignancies. Blood 2017, 130, 2815.
- Li, Z.; Krippendorff, B.-F.; Sharma, S.; Walz, A.C.; Lavé, T.; Shah, D.K. Influence of molecular size on tissue distribution of antibody fragments. mAbs 2016, 8, 113–119.
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 242, 423–426.
- Gaissmaier, L.; Christopoulos, P. Immune Modulation in Lung Cancer: Current Concepts and Future Strategies. Respiration 2020, 1–27.
More
