Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Vivi Li and Version 2 by Mario Andrea Marchisio.
Among noncoding RNA sequences, riboswitches and ribozymes have attracted the attention of the synthetic biology community as circuit components for translation regulation. When fused to aptamer sequences, ribozymes and riboswitches are enabled to interact with chemicals. Therefore, protein synthesis can be controlled at the mRNA level without the need for transcription factors. Potentially, the use of chemical-responsive ribozymes/riboswitches would drastically simplify the design of genetic circuits.
- S. cerevisiae
- aptamers
- riboswitches
- ribozymes
- synthetic biology
1. Introduction
RNA is a polymeric biomacromolecule that carries out crucial functions in diverse biological systems. Since it was discovered, the messenger RNA (mRNA) has shown its fundamental role in gene expression as the intermediate step from DNA to proteins in the central dogma. Further, noncoding RNA elements are ubiquitous both in prokaryotic and eukaryotic cells. Over the last two decades, with the advent of synthetic biology, non-coding RNAs have been largely used to control protein synthesis within genetic circuits [1]. To this aim, riboswitches and ribozymes have proved to be powerful solutions due to their capability to interact with chemicals, i.e., circuit inputs [2].
Riboswitches are regions of the mRNA that contain specific evolutionarily conserved ligand-binding domains (sensors) along with a variable sequence, termed the expression platform, which enables regulation of the downstream coding sequence (CDS) [3]. They were discovered in 2002 in bacteria [4][5][6] and later exploited in numerous species for tuning RNA stability [7], transcription [8], translation [9], and RNA splicing [10]. Riboswitch sensors are RNA aptamers, i.e., hairpin structures that bind small molecules with high affinity and specificity. Usually, synthetic aptamers are produced through in vitro or in vivo selection—systematic evolution of ligands by exponential enrichment (SELEX) [11][12]. Artificial aptamers have been selected to bind different bio-targets such as dyes, proteins, aromatic small molecules, and antibiotics [13]. Upon ligand binding, aptamers undergo conformational changes that have an impact on the tertiary structure (folding) of the mRNA chain where the riboswitch is included. This explains why natural and synthetic riboswitches are a means of controlling translation initiation.
Ribozymes—whose existence has been known since the 1980s of the last century [14]—are functional RNA molecules able to catalyze biochemical reactions such as the cleavage and the ligation of phosphodiester and peptide bonds [15][16][17]. Therefore, in their activity they resemble enzyme proteins. However, ribozymes have unique characteristics and advantages over proteinic enzymes: (1) They are functional RNA transcripts, i.e., they do not basically demand any particular post-transcriptional modifications; (2) the expression level of functional RNA transcripts is more controllable and predictable than that of proteins because of the absence of the translational step; (3) transcription consumes less energy and resources than translation; (4) manipulation of ribozyme functionality is relatively easy compared to that of polypeptides; and (5) the activity and the stability of ribozymes can be tuned via simple modifications [18]. The catalytic activities of ribozymes are generally categorized into three groups: Cleavage, splicing, and other functions [18]. Cleavage is the most widespread task among ribozymes. We find it in self-cleaving hammerhead ribozymes (HHR) [19], simple hairpins [20], Varkud satellites (VS) [21], hepatitis delta virus (HDV) ribozymes [22], glucosamine-6-phosphate synthase (glmS) ribozymes [23], and twister ribozymes [24]. Aptamers fused to natural, self-catalytic RNA structures are, in general, referred to as aptazymes [25][26]. They have broadened the potential of riboswitches/ribozymes for predictable gene regulation and have been widely engineered [27].
2. Ligand-Responsive Aptamers in S. cerevisiae
Most of the ribozymes and riboswitches that have shown high functionality in S. cerevisiae respond to well-tolerated drugs, such as theophylline, tetracycline, and aminoglycosides. They are thoroughly described in the following. Examples of other RNA structures sensing different chemicals are given as well, at the end of this section.
2.1. Theophylline Aptamers
Theophylline (1,3-dimethylxanthine—see Figure 1a), a common medicine for the treatment of chronic asthma, is a natural product extracted from the leaves of plants such as Camellia sinensis (tea plant) and Theobroma cacao. In 1994, Jenison et al. [28] used SELEX to isolate RNA molecules with high affinity for theophylline. From the pool of selected molecules, TCT8-4 RNA was truncated into the mTCT8-4 aptamer that is now simply referred to as the canonical theophylline aptamer [29]. This sequence shortening increased the affinity of the RNA for the drug, as the dissociation constant KD decreased from 0.6 to 0.1 µM [28]. The theophylline aptamer (Figure 1b) is over 30-nt long and includes a core made of fifteen conserved residues, organized into two internal loops, which is required for theophylline binding [28][30][31]. In particular, C27 nucleotide is the key residue in recognizing theophylline and discriminating against caffeine [31]. The aptamer manifests a structural change upon chemical binding (conformational capture, or selection, and mechanism). Initially, this RNA structure was utilized to build allosteric ribozymes such as the above-mentioned aptazymes. In 2005, Bayer and Smolke [32] developed a class of small, modular trans-acting RNAs, termed antiswitches, which regulated gene expression in a tunable ligand-dependent manner. They fused theophylline (mainly) and tetracycline aptamers to an antisense RNA sequence that was designed to match a fifteen-nucleotide region near the start codon of the target mRNA (encoding for the green fluorescence protein—GFP) (Figure 1c). Implementation of antiswitches in S. cerevisiae determined a sharp reduction (OFF switch) or increase (ON switch)—about 90% in both cases—in GFP expression in the presence of 1 to 10 mM of theophylline. This work represented the first successful application of theophylline-dependent riboswitches in eukaryotic cells.
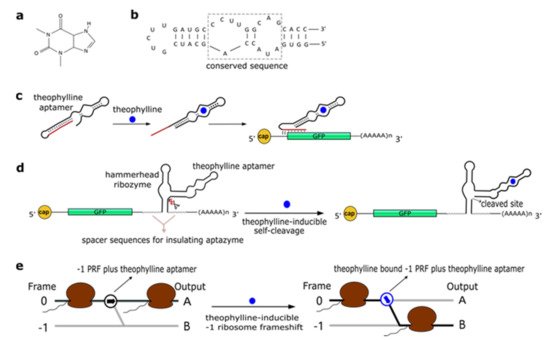
Figure 1. The theophylline aptamer. (a) Theophylline structure (PubChem CID 2153). (b) The secondary structure of the theophylline aptamer. (c) The theophylline-responsive antiswitch as a means to control gene expression [32]. The RNA sequence complementary to the target green fluorescence protein (GFP) transcript is highlighted in red. (d) Fusion of a theophylline aptamer to the stem II of a hammerhead ribozyme. The resulting aptazyme undergoes autocleavage in the presence of 5 µM of theophylline causing an over 15-fold decrease in fluorescence expression. (e) Theophylline-induced translational frame shift. Here, the theophylline aptamer is fused to a ‒1 programmed ribosomal frameshifting (PRF) RNA structure [33].
In a later work, Win and Smolke [34] modified an HHR with the insertion of either a theophylline or a tetracycline aptamer at the stem II by means of a communication module (referred to as transmitter). Chemical binding to the aptamer triggered the tertiary interactions between loop I and II, necessary either to induce or block self-cleavage. Stem III was used to integrate the synthetic ribozyme in the 3′UTR (untranslated region) of the target mRNA (Figure 1d). This “RNA-based gene-regulatory platform” showed, in S. cerevisiae, its utility in the control of cell growth and the detection of metabolites such as xanthine. This design was subsequently modified with the usage of a transmitter to link an aptamer to either stem I/stem II, or another aptamer. Single-input complex devices and two-input Boolean gates (such as AND, NAND, NOR, and OR) sensing theophylline (5–10 mM) and/or tetracycline (0.25–0.5 mM) were engineered by means of two modified HHRs or a single one carrying an aptamer at both stem I and stem II or two aptamers in series at stem II. A remarkable ~14-fold gain in fluorescence expression was achieved by the AND gate [35]. A single HHR modified with the addition of only one theophylline aptamer (tetracycline in few variants) was then used to establish a two-color fluorescence-activated cell sorting (FACS)-based screening approach [36]. This modular design for theophylline-responsive ribozymes was, furthermore, adopted to implement a novel screening system for enzyme activity. By exploiting the fact that theophylline is produced via caffeine demethylation, the activity of a caffeine demethylase in S. cerevisiae could be assessed by the fluorescence level triggered by an ON switch sensing the amount of theophylline (up to 140 µM) derived from (up to 1 mM) caffeine transformation. This screening process permitted us to find out mutations that increased, considerably, both the activity and the selectivity of caffeine demethylase [37].
A peculiar implementation of RNA-based Boolean gates and an apoptosis controller was presented by Anzalone et al. [33] who engineered ligand-responsive ‒1 programmed ribosomal frameshifting (PRF) elements by coupling them with either theophylline or neomycin aptamers. Eukaryotic ‒1 PRF signals, present inside mRNA transcripts, are made of two main features: (1) The slippery site, where the frameshift takes place, which corresponds to two homopolymeric triplets characterized by the sequence XXXYYYZ (X denoting any nucleotides; Y standing for A or U; and Z meaning A, C or U); and (2) a stimulatory RNA structure (a hairpin or a pseudoknot) located downstream of the heptanucleotide. When, during the elongation phase, ribosomes meet a ‒1 PRF signal along the mRNA, a fraction of them slip back by a single nucleotide, shifting translation to the ‒1 reading frame, with a consequent change in the polypeptide downstream of the ‒1 PRF signal (Figure 1e) [38]. Anzalone and co-authors combined optimized ‒1 PRF RNA structures with aptamers either to prevent (OFF switch) or enhance (ON switch) the frameshift in translation in the presence of the aptamer-binding ligands. In both configurations, theophylline aptamers slightly outperformed neomycin aptamers. In particular, 40 mM theophylline led either to a 7.0-fold increase or a 5.9-fold decrease of the translation frameshift, whereas 550 µM neomycin allowed only either a 5.0-fold enhancement or a 4.2-fold reduction of the same event. Two OFF switches led to a NOR gate implementation, whereas an AND gate demanded two ON switches. A theophylline ON switch coupled to a neomycin OFF switch was, in contrast, at the basis of the apoptosis controller. Each synthetic circuit was shown to work with high efficiency, with the apoptosis controller allowed us to register an over 300-fold decrease in cell viability in the presence of both inputs.
2.2. Tetracycline Aptamers
Promoters activated or repressed by tetracycline (tc—see Figure 2a) have been used in yeast bio-engineering for over twenty years [39][40] since tc is an easy-to-handle antibiotic that does not interfere severely with yeast cellular metabolism [41].
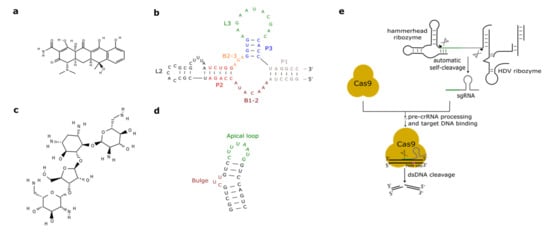
Figure 2. Antibiotic-responsive aptamers and the ribozyme-gRNA-ribozyme (RGR) cassette. (a) Tetracycline structure (PubChem CID 54675776). (b) The tetracycline aptamer. (c) Neomycin structure (PubChem CID 8378). (d) The neomycin aptamer. The main structural features of the two RNA molecules are here highlighted. (e) RGR cassette for the expression of single-guide RNAs. Upon autocleavage of the HH and the hepatitis delta virus (HDV) ribozymes, the single guide RNA is released. sgRNA binds Cas9 and brings it to the target DNA that is finally cut. The green line denotes the spacer, whereas the gray hairpin represents the direct repeat.
As we have seen in the previous section, the tc aptamer has been used in several works together with the theophylline aptamer. The tetracycline aptamer consists of three stems (P1–P3), two single-stranded regions (the bulges, B1-2 and B2-3), and the loops L2 and L3 (see Figure 2b). Portions of both bulges and the L3 loop are involved in tc binding [42] that takes place at a very high affinity (KD equals 0.8 nM [43]). In 2003, Suess et al. [44] reported the application of a tetracycline-binding aptamer to the control of gene expression in S. cerevisiae. Green fluorescence was measured after placing the tc-responsive riboswitch in the 5′UTR of the strong ADH1 promoter (pADH1), either close to the fluorescent protein start codon or in a cap-proximal location. The latter position appeared more effective in repression of translation (6-fold in the presence of tetracycline). Interestingly, stabilization of the aptamer structure led to an increased ON-to-OFF ratio but lowered basal fluorescence expression (i.e., in the absence of tetracycline). In another work published in 2003, the tetracycline riboswitch was shown to lead, in the presence of the antibiotic, to a 9-fold downregulation in luciferase synthesis from the constitutive TEF1 promoter [45]. 5′UTRs containing a variable number (one to three) of tetracycline aptamers were analyzed in [46]. Initially, each configuration was placed between the ADH1 promoter and the GFP gene. As already experienced in [44][45], riboswitches lowered the cell fluorescence level considerably when already in the absence of tetracycline. However, in the presence of 250 µM of the chemical, two and three aptamers caused an almost complete loss of fluorescence (1.1% and 0.6% of the fluorescence produced by pADH1 without aptamers) with a 21-fold and 37-fold repression factor compared to tetracycline-untreated cells. These riboswitch-containing 5′UTRs, preceded by either pADH1 or the GPD promoter, were then adopted to construct universal insertion cassettes that could be integrated, via homologous recombination, into the yeast genome. Five endogenous genes (NEP1, NOP8, NOP14, PGI1, and SEC1) were completely switched OFF upon induction with at most 500 µM of tetracycline. NOP8 and SEC1 had never been regulated previously. More recently, the tandem configuration of this riboswitch was optimized in silico via machine learning techniques. In vivo, the newly designed 5′UTR sequence guaranteed a 40-fold repression of fluorescence in the presence of 250 µM tetracycline [42].
One or two tetracycline-responsive riboswitches were also shown to be effective in controlling pre-mRNA splicing when placed within an intron nearby the 5′ splice site (SS). The most efficient regulation was obtained by burying the 5′ SS into the P1 stem at the basis of the tetracycline aptamer. Upon insertion of the modified intron along the sequence of GFP, fluorescence was emitted by the cells only in the absence of tetracycline, i.e., when the mRNA was spliced successfully. In contrast, in the presence of tetracycline a drastic reduction in cell fluorescence level was observed [47].
In a different approach to control gene expression, the tetracycline aptamer was fused to the full-length HHR N79 from Schistosoma mansoni via a linker region optimized through 11 rounds of in vitro selection (SELEX). Synthetic ribozymes completely self-cleaved in the presence of 1 µM of tetracycline, whereas self-cleavage was inhibited up to 333-fold in the absence of the antibiotic [48].
2.3. Aminoglycosides-Responsive Aptamers
Aminoglycosides are positively charged antibiotics characterized by a high affinity towards RNA sequences [49][50][51]. Among them in the 1990s, neomycin was shown to have a potent inhibitory action on different kinds of ribozymes [52][53]. Hence, Weigand et al. [54] employed neomycin (Figure 2c) to develop a two-stage method—in vitro selection followed by in vivo (yeast) screening—to identify novel ligand-binding riboswitches. With this procedure, several new artificial aptamers responding to neomycin were engineered. The most performant one, termed M4, induced a 7.5-fold fluorescence repression in the presence of 100 µM neomycin. Another neomycin-binding aptamer, N1 (see Figure 2d), was subsequently deeply studied and its interactions with different aminoglycosides (such as ribostamycin, tobramycin, and paromomycin) were clarified [55][56][57][58][59].
Klauser et al. [60][61] introduced a new design for synthetic ligand-responsive ribozymes. The main idea was to preserve, as far as possible, the structure of a native HHR to which either a theophylline or a neomycin (N1) aptamer was attached. This was achieved by placing the aptamer at the 5′ end (stem III) of a type 3 S. mansoni HHR. Neomycin-responsive ribozymes, engineered in this way, were inserted in the 3′UTR of the gal4 gene. Hence, gal4 expression was maximal in the absence of neomycin. The Gal4 activator induced the synthesis of lacZ such that the ribozyme performance was assessed, indirectly, via beta-galactosidase assay. This represented a new protocol for the in vivo selection of neomycin-sensing ribozymes. The best one reported in this work showed a ~25-fold reporter downregulation in the presence of 100 µg/mL neomycin. A similar design was followed, in a later work by the same group [62], for the engineering of a neomycin-responsive riboswitch, based on an inactive env-9 twister ribozyme, which produced a ~10-fold downregulation in the lacZ gene expression.
In a recent work, Sack et al. [63] presented a new design for the neomycin ribozyme where a neomycin aptamer (M4 or M7 [54]) was placed at stem I of the S. mansoni HHR. The connection between aptamer and stem I was realized through an addressable three-way junction [64], whose flexibility was increased with the addition of a three-nucleotide bulge (UAU). The synthetic ribozyme was inserted in the 3′UTR of the gfp gene such that its activity was quantified, directly, by measuring cell fluorescence. Libraries of artificial ribozymes, obtained by randomizing, first, the three-way junction and, then, the three-nucleotide bulge, were screened in the presence and absence of, usually, 100 µg/mL of neomycin. Differently from [65], both OFF and ON switches were found, i.e., the binding of neomycin, in some cases, made the HHR fold into an inactive conformation and cause an increase in green protein synthesis. ON switches achieved a ~2-fold fluorescence increase, whereas OFF switches caused a ~3-fold decrease in fluorescence.
A new synthetic riboswitch responding to paromomycin [66] was engineered after screening a library of 1015 RNA sequences via the Capture-SELEX method [67]. With respect to SELEX, Capture-SELEX allows, in vitro, a first selection of aptamers that undergo structural modification upon binding the chosen ligand. This lowers considerably the number of riboswitches (or ribozymes) to be tested, subsequently, for their in vivo functionality. The synthetic PARO riboswitch is 44-nt long and consists of two hairpins (P1 and P2) with the docking site located on P2. It binds paromomycin with high affinity (KD = 20 nM) and allows an 8.5-fold translation downregulation to be registered when exposed to 250 µM of the antibiotic. On it, a working NOR gate was engineered by replacing P1 with the neomycin aptamer. In the presence of 250 µM of both inputs, the logic gate gave a 13.8-fold repression factor.
2.4. Other Aptamers Used in S. cerevisiae
As we have seen above, antibiotics such as tetracycline and aminoglycosides have been largely used for the engineering of novel aptamers. Groher et al. [68] realized a synthetic riboswitch responding to a new kind of antibiotic, the fluoroquinolone ciprofloxacin. Tested on GFP expression, the riboswitch showed a 7.5-fold fluorescence downregulation and a good affinity towards its ligand (KD = 64.2 nM).
In 2001, Grate and Wilson [69] described a method to influence S. cerevisiae cell cycle via a dye-responsive aptamer placed in the 5′UTR of the CLB2 gene. Upon binding tetramethylrosamine (a malachite green analog), the aptamer underwent a drastic structural change that reduced CLB2 translation in a considerable way. As a consequence, cell growth slowed down and elongated buds appeared during cell division.
More recently, a novel kind of aptamer, indirectly activated by light, was realized by Lotz et al. [70]. This RNA structure binds molecules of azoCm, i.e., the trans photoisomer of azobenzene modified with chloramphenicol. Upon irradiation with 360-nm-wavelength light, azoCm falls into a configuration (the cis photoisomer) that is unable to bind the RNA aptamer. The RNA-binding trans photoisomer is re-established by exposing the cis photoisomer to light at 420-nm wavelength. Hence, light activation is conferred to the azoCm aptamer by the peculiar characteristics of its ligand.
After describing many synthetic aptamers, we shall mention the thiamine pyrophosphate (TPP) riboswitches, i.e., the only natural eukaryotic riboswitches discovered so far. They were found—usually in introns, as splicing regulators—in the genome of plants [71], filamentous fungi [72], algae, and marine phytoplankton [73]. Donovan et al. [74] showed that TPP riboswitches are present in some budding yeast species, such as Candida parapsilosis, and are functional in S. cerevisiae. Once placed on an intron inside a reporter protein gene, the TPP riboswitch allowed splicing and, therefore, fluorescence expression only in the absence of thiamine, whereas the pre-mature mRNA was not spliced in the presence of 10 µM thiamine.
An overview of the performance of the aptamers described in this section is given in Table 1.
Table 1. Aptamer performance overview. The effect on gene expression due to the chemical-responding aptamers described in this work are here summarized.
Aptamer performance overview. The effect on gene expression due to the chemical-responding aptamers described in this work are here summarized.
| RNA Structure | Kd | Additional RNA | Action Triggered by the Chemicals | Performance | Reference(s) |
|---|---|---|---|---|---|
| Theophylline aptamer mTCT8-4 | 0.1 μM | Short antisense RNA | Increase/decrease in gene expression upon binding near the START codon | 90% reduction (OFF switch) or increase (ON switch) in fluorescence expression (1 to 10 mM of theophylline) | [32] |
| HHR | Induction/inhibition of ribozyme self-cleavage | 14-fold increase in fluorescence expression from an AND gate (5–10 mM theophylline and 0.25–0.5 mM tetracycline) | [33,34] | ||
| - | Translational frame shift (-1 PRF) | 7.0-fold increase or 5.9-fold decrease in the translation frame shift (40 mM theophylline) | [38] | ||
| Tetracycline aptamer | 0.8 nM | - | Translation inhibition upon placement on the 5’ UTR of gfp | Single aptamer: 6-to 9-fold fluorescence repression; 2 and 3 aptamers: 21-fold and 37-fold fluorescence repression, respectively. In every case, 250 μM tetracycline were used | [44,45] |
| Intron | Pre-mRNA splicing | Unquantified fluorescence reduction | [47] | ||
| HHR | Ribozyme self-cleavage | Complete self-cleaved (1 μM tetracycline) | [48] | ||
| Neomycin aptamer | - | - | Translational frame shift (-1 PRF) | 5.0-fold enhancement or 4.2-fold reduction in the translation frame shift (550 µM neomycin) | [37] |
| Neomycin aptamer N1 Neomycin aptamer N1-based riboswitch |
- - |
S. mansoni HHR | Translation inhibition upon insertion on the 3’UTR of gal4 | Around 25-fold lacZ expression downregulation (100 μg/mL) | [60,61] |
| Inactive env-9 twister ribozyme | Translation inhibition upon insertion on the 3’UTR of gal4 | About 10-fold decrease in lacZ expression | [62] | ||
| Neomycin aptamer M4 or M7 | - | S. mansoni HRR | Translation inhibition upon insertion on the 3’UTR of gfp | Around 2-fold fluorescence upregulation (ON switch) and 3-fold fluorescence downregulation (OFF switch) (100 μg/mL neomycin) | [64] |
| Neomycin aptamer M4 | - | - | Translation inhibition | 7.5-fold fluorescence repression (100 µM neomycin) | [54] |
| PARO riboswitch (paromomycin) |
20 nM | - | Translation inhibition | 8.5-fold decrease in gene expression (250 μM paromomycin) | [66,67] |
| 13.8-fold decrease in gene expression from a NOR gate (250 μM of both paromomycin and neomycin) | |||||
| fluoroquinolone ciprofloxacin riboswitch | 64.2 nM | - | Translation inhibition | 7.5-fold fluorescence downregulation | [68] |
| Tetra- methylrosamine aptamer |
- | - | Translation inhibition upon placement on the 5’ UTR of CLB2 | Reduction in cell growth | [69] |
| azoCm aptamer | - | - | Configurational change | Unquantified control of gene expression | [70] |
| TPP (thiamine pyrophosphate) riboswitch | - | Intron | Splicing inhibition | pre-mRNA is not spliced in the presence of 10 µM thiamine | [74] |
References
- Benenson, Y. Synthetic biology with RNA: Progress report. Curr. Opin. Chem. Biol. 2012, 16, 278–284.
- Marchisio, M.A.; Stelling, J. Automatic design of digital synthetic gene circuits. PLoS Comput. Biol. 2011, 7, e1001083.
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24.
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756.
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043–1049.
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956.
- Schroeder, K.T.; Daldrop, P.; Lilley, D.M.J. RNA Tertiary Interactions in a Riboswitch Stabilize the Structure of a Kink Turn. Structure 2011, 19, 1233–1240.
- Hollands, K.; Proshkin, S.; Sklyarova, S.; Epshtein, V.; Mironov, A.; Nudler, E.; Groisman, E.A. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. USA 2012, 109, 5376–5381.
- Breaker, R.R. Riboswitches and Translation Control. Cold Spring Harb. Perspect. Biol. 2018, 10.
- Kim, D.S.; Gusti, V.; Pillai, S.G.; Gaur, R.K. An artificial riboswitch for controlling pre-mRNA splicing. RNA 2005, 11, 1667–1677.
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822.
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510.
- Hermann, T.; Patel, D.J. Biochemistry—Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825.
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157.
- Baskerville, S.; Bartel, D.P. A ribozyme that ligates RNA to protein. Proc. Natl. Acad. Sci. USA 2002, 99, 9154–9159.
- Doudna, J.A.; Cech, T.R. The chemical repertoire of natural ribozymes. Nature 2002, 418, 222–228.
- Serganov, A.; Keiper, S.; Malinina, L.; Tereshko, V.; Skripkin, E.; Hobartner, C.; Polonskaia, A.; Phan, A.T.; Wombacher, R.; Micura, R.; et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 2005, 12, 218–224.
- Park, S.V.; Yang, J.S.; Jo, H.; Kang, B.; Oh, S.S.; Jung, G.Y. Catalytic RNA, ribozyme, and its applications in synthetic biology. Biotechnol. Adv. 2019, 37, 107452.
- O’Rourke, S.M.; Scott, W.G. Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme. Prog. Mol. Biol. Transl. Sci. 2018, 159, 177–202.
- Ren, A.; Micura, R.; Patel, D.J. Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr. Opin. Chem. Biol. 2017, 41, 71–83.
- Lacroix-Labonte, J.; Girard, N.; Dagenais, P.; Legault, P. Rational engineering of the Neurospora VS ribozyme to allow substrate recognition via different kissing-loop interactions. Nucleic Acids Res. 2016, 44, 6924–6934.
- Been, M.D.; Wickham, G.S. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 1997, 247, 741–753.
- Savinov, A.; Block, S.M. Self-cleavage of the glmS ribozyme core is controlled by a fragile folding element. Proc. Natl. Acad. Sci. USA 2018, 115, 11976–11981.
- Roth, A.; Weinberg, Z.; Chen, A.G.; Kim, P.B.; Ames, T.D.; Breaker, R.R. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014, 10, 56–60.
- Tang, J.; Breaker, R.R. Rational design of allosteric ribozymes. Chem. Biol. 1997, 4, 453–459.
- Soukup, G.A.; Breaker, R.R. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. USA 1999, 96, 3584–3589.
- Zhong, G.; Wang, H.; Bailey, C.C.; Gao, G.; Farzan, M. Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. Elife 2016, 5.
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429.
- Wrist, A.; Sun, W.; Summers, R.M. The Theophylline Aptamer: 25 Years as an Important Tool in Cellular Engineering Research. ACS Synth. Biol. 2020, 9, 682–697.
- Lee, S.W.; Zhao, L.; Pardi, A.; Xia, T. Ultrafast Dynamics Show That the Theophylline and 3-Methylxanthine Aptamers Employ a Conformational Capture Mechanism for Binding Their Ligands. Biochemistry 2010, 49, 2943–2951.
- Zimmermann, G.R.; Jenison, R.D.; Wick, C.L.; Simorre, J.P.; Pardi, A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997, 4, 644–649.
- Bayer, T.S.; Smolke, C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005, 23, 337–343.
- Anzalone, A.V.; Lin, A.J.; Zairis, S.; Rabadan, R.; Cornish, V.W. Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat. Methods 2016, 13, 453–458.
- Win, M.N.; Smolke, C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA 2007, 104, 14283–14288.
- Win, M.N.; Smolke, C.D. Higher-order cellular information processing with synthetic RNA devices. Science 2008, 322, 456–460.
- Liang, J.C.; Chang, A.L.; Kennedy, A.B.; Smolke, C.D. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012, 40, e154.
- Michener, J.K.; Smolke, C.D. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab. Eng. 2012, 14, 306–316.
- Brierley, I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995, 76 Pt 8, 1885–1892.
- Zilio, N.; Wehrkamp-Richter, S.; Boddy, M.N. A new versatile system for rapid control of gene expression in the fission yeast Schizosaccharomyces pombe. Yeast 2012, 29, 425–434.
- Belli, G.; Gari, E.; Piedrafita, L.; Aldea, N.; Herrero, E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998, 26, 942–947.
- Wishart, J.A.; Hayes, A.; Wardleworth, L.; Zhang, N.S.; Oliver, S.G. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast 2005, 22, 565–569.
- Groher, A.C.; Jager, S.; Schneider, C.; Groher, F.; Hamacher, K.; Suess, B. Tuning the Performance of Synthetic Riboswitches using Machine Learning. ACS Synth. Biol. 2019, 8, 34–44.
- Foerster, U.; Weigand, J.E.; Trojanowski, P.; Suess, B.; Wachtveitl, J. Conformational dynamics of the tetracycline-binding aptamer. Nucleic Acids Res. 2012, 40, 1807–1817.
- Suess, B.; Hanson, S.; Berens, C.; Fink, B.; Schroeder, R.; Hillen, W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003, 31, 1853–1858.
- Hanson, S.; Berthelot, K.; Fink, B.; McCarthy, J.E.; Suess, B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003, 49, 1627–1637.
- Kotter, P.; Weigand, J.E.; Meyer, B.; Entian, K.D.; Suess, B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009, 37, e120.
- Weigand, J.E.; Suess, B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007, 35, 4179–4185.
- Wittmann, A.; Suess, B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. Biosyst. 2011, 7, 2419–2427.
- Walter, F.; Vicens, Q.; Westhof, E. Aminoglycoside—RNA interactions. Curr. Opin. Chem. Biol. 1999, 3, 694–704.
- Schroeder, R.; Waldsich, C.; Wank, H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000, 19, 1–9.
- Zhao, F.; Zhao, Q.; Blount, K.F.; Han, Q.; Tor, Y.; Hermann, T. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew. Chem. Int. Ed. 2005, 44, 5329–5334.
- Stage, T.K.; Hertel, K.J.; Uhlenbeck, O.C. Inhibition of the hammerhead ribozyme by neomycin. RNA 1995, 1, 95–101.
- Von Ahsen, U.; Davies, J.; Schroeder, R. Antibiotic inhibition of group I ribozyme function. Nature 1991, 353, 368–370.
- Weigand, J.E.; Sanchez, M.; Gunnesch, E.B.; Zeiher, S.; Schroeder, R.; Suess, B. Screening for engineered neomycin riboswitches that control translation initiation. RNA 2008, 14, 89–97.
- Weigand, J.E.; Schmidtke, S.R.; Will, T.J.; Duchardt-Ferner, E.; Hammann, C.; Woehnert, J.; Suess, B. Mechanistic insights into an engineered riboswitch: A switching element which confers riboswitch activity. Nucleic Acids Res. 2011, 39, 3363–3372.
- Duchardt-Ferner, E.; Weigand, J.E.; Ohlenschlaeger, O.; Schtnidtke, S.R.; Suess, B.; Woehnert, J. Highly Modular Structure and Ligand Binding by Conformational Capture in a Minimalistic Riboswitch. Angew. Chem. Int. Ed. 2010, 49, 6216–6219.
- Schmidtke, S.R.; Duchardt-Ferner, E.; Weigand, J.E.; Suess, B.; Woehnert, J. NMR resonance assignments of an engineered neomycin-sensing riboswitch RNA bound to ribostamycin and tobramycin. Biomol. Nmr Assign. 2010, 4, 115–118.
- Krstic, I.; Frolow, O.; Sezer, D.; Endeward, B.; Weigand, J.E.; Suess, B.; Engels, J.W.; Prisner, T.F. PELDOR spectroscopy reveals preorganization of the neomycin-responsive riboswitch tertiary structure. J. Am. Chem. Soc. 2010, 132, 1454–1455.
- Kulik, M.; Mori, T.; Sugita, Y.; Trylska, J. Molecular mechanisms for dynamic regulation of N1 riboswitch by aminoglycosides. Nucleic Acids Res. 2018, 46, 9960–9970.
- Klauser, B.; Atanasov, J.; Siewert, L.K.; Hartig, J.S. Ribozyme-based aminoglycoside switches of gene expression engineered by genetic selection in S. cerevisiae. ACS Synth. Biol. 2015, 4, 516–525.
- Klauser, B.; Rehm, C.; Summerer, D.; Hartig, J.S. Engineering of Ribozyme-Based Aminoglycoside Switches of Gene Expression by In Vivo Genetic Selection in Saccharomyces cerevisiae. Methods Enzymol. 2015, 550, 301–320.
- Felletti, M.; Stifel, J.; Wurmthaler, L.A.; Geiger, S.; Hartig, J.S. Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nat. Commun. 2016, 7, 12834.
- Sack, M.; Stifel, J.; Kreft, S.G.; Deuerling, E.; Hartig, J.S. Neomycin-dependent hammerhead ribozymes for the direct control of gene expression in Saccharomyces cerevisiae. Methods 2019, 161, 35–40.
- Wieland, M.; Gfell, M.; Hartig, J.S. Expanded hammerhead ribozymes containing addressable three-way junctions. RNA 2009, 15, 968–976.
- Murphy, G.J.; Mostoslavsky, G.; Kotton, D.N.; Mulligan, R.C. Exogenous control of mammalian gene expression via modulation of translational termination. Nat. Med. 2006, 12, 1093–1099.
- Boussebayle, A.; Torka, D.; Ollivaud, S.; Braun, J.; Bofill-Bosch, C.; Dombrowski, M.; Groher, F.; Hamacher, K.; Suess, B. Next-level riboswitch development-implementation of Capture-SELEX facilitates identification of a new synthetic riboswitch. Nucleic Acids Res. 2019, 47, 4883–4895.
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal. Methods Chem. 2012, 2012, 415697.
- Groher, F.; Bofill-Bosch, C.; Schneider, C.; Braun, J.; Jager, S.; Geissler, K.; Hamacher, K.; Suess, B. Riboswitching with ciprofloxacin-development and characterization of a novel RNA regulator. Nucleic Acids Res. 2018, 46, 2121–2132.
- Grate, D.; Wilson, C. Inducible regulation of the S-cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001, 9, 2565–2570.
- Lotz, T.S.; Halbritter, T.; Kaiser, C.; Rudolph, M.M.; Kraus, L.; Groher, F.; Steinwand, S.; Wachtveitl, J.; Heckel, A.; Suess, B. A light-responsive RNA aptamer for an azobenzene derivative. Nucleic Acids Res. 2019, 47, 2029–2040.
- Wachter, A.; Tunc-Ozdemir, M.; Grove, B.C.; Green, P.J.; Shintani, D.K.; Breaker, R.R. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant. Cell 2007, 19, 3437–3450.
- Cheah, M.T.; Wachter, A.; Sudarsan, N.; Breaker, R.R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 2007, 447, 497–500.
- McRose, D.; Guo, J.; Monier, A.; Sudek, S.; Wilken, S.; Yan, S.; Mock, T.; Archibald, J.M.; Begley, T.P.; Reyes-Prieto, A.; et al. Alternatives to vitamin B-1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J. 2014, 8, 2517–2529.
- Donovan, P.D.; Holland, L.M.; Lombardi, L.; Coughlan, A.Y.; Higgins, D.G.; Wolfe, K.H.; Butler, G. TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida. PLoS Genet. 2018, 14, e1007429.
More
