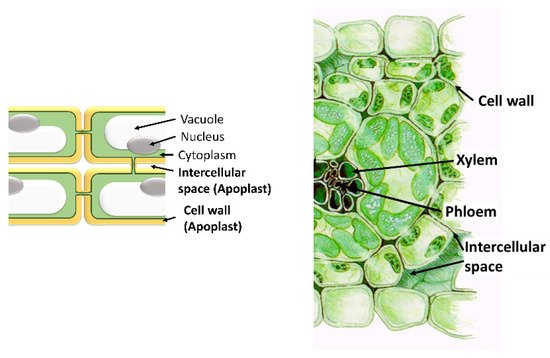
The apoplast comprises the intercellular space, the cell walls, and the xylem. Important functions for the plant, such as nutrient and water transport, cellulose synthesis, and the synthesis of molecules involved in plant defense against both biotic and abiotic stresses, take place in it. The most important molecules are ROS, antioxidants, proteins, and hormones. Even though only a small quantity of ROS is localized within the apoplast, apoplastic ROS have an important role in plant development and plant responses to various stress conditions. In the apoplast, like in the intracellular cell compartments, a specific set of antioxidants can be found that can detoxify the different types of ROS produced in it. These scavenging ROS components confer stress tolerance and avoid cellular damage. Moreover, the production and accumulation of proteins and peptides in the apoplast take place in response to various stresses. Hormones are also present in the apoplast where they perform important functions. In addition, the apoplast is also the space where microbe-associated molecular Patterns (MAMPs) are secreted by pathogens. In summary, the diversity of molecules found in the apoplast highlights its importance in the survival of plant cells.
- apoplast
- plant defense
- ROS
- antioxidants
- proteins
- peptides
- hormones
- MAMPs
1. Introduction
2. Antioxidants in the Apoplast
As mentioned above, ROS can be at the same time beneficial and deleterious for the plant, since they can act as secondary messengers in different physiological processes [65][23]. However, they can also induce oxidative damage under several environmental stress conditions, such as salinity, drought, cold, heavy metals, and UV irradiation. In the latter case, ROS accumulation may cause many cellular damages that consist of degradation of several biomolecules such as pigments, proteins, lipids, carbohydrates, and DNA, which finally leads to PCD. It was revealed that a high concentration of ROS has a toxic role in plant cells, so the actions of ROS scavengers and antioxidant enzymes are required to avoid its toxicity [65][23]. The components of the antioxidant machinery can be broadly divided into enzymatic and non-enzymatic. The relevant ROS-scavenging enzymes are superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), and glutathione reductase (GR) (Table 1). Moreover, the non-enzymatic antioxidants such as ascorbate, glutathione, proline, phenolic compounds, and polyamines are also required to avoid cell toxicity [66][24] (Table 2). Superoxide dismutases (SOD) are enzymes that convert O2•− into H2O2, which is less harmful to the plant. Therefore, they are considered as the first line of detoxification of ROS. SODs are classified according to the metal cofactor used by the enzyme, as manganese (Mn-SOD), iron (Fe-SOD), and copper–zinc (Cu/Zn-SOD) dependent, although Cu/ZnSOD was identified as the most active apoplastic isoform. Furthermore, many studies have reported that these enzymes confer resistance to abiotic stress. These studies highlight the fact that the tolerance level of plants is positively correlated with SOD activity and with the number of SOD isoforms. SOD is up-regulated in many plant species by various types of abiotic stress-inducing factors, such as drought, salt, and heavy metals, in a large number of crops, such as, tomato, wheat, barley, and citrus [67][25]. Related to this, it was shown that apROS production in wheat root decreased upon a copper treatment, which was correlated with an increase in SOD activity [68][26]. Moreover, Garcia et al. [69][27] found a positive correlation between SOD activity and salt-tolerance when studying the response of the apoplastic antioxidant systems in root and leaf tissues from a salt-sensitive and a salt-resistant onion genotype in response to salinity. Furthermore, increased SOD activity was also observed in response to biotic stress. Vanacker et al. [70][28] showed that the activities of several antioxidant enzymes such as SOD, CAT, APX, DHAR, MDHAR, and GR were induced in the apoplasts of barley and oat leaves 24 h after inoculation with the biotrophic fungal pathogen Blumeria graminis [71][29]. Furthermore, Trichoderma harzianum inoculated sunflower plants were more resistant to Rhizoctonia solani. This resistance was accompanied by an increase in SOD activity in root apoplast seven days post-inoculation [72][30].| ENZYMATIC ANTIOXIDANTS | ||||
|---|---|---|---|---|
| Enzyme | Chemical Reaction | Involved in | Cellular Location | Ref. |
| NON-ENZYMATIC ANTIOXIDANTS | ||||||
|---|---|---|---|---|---|---|
| Enzyme | Functions | Location | Ref. | |||
| Superoxide Dismutase (SOD) | O2•−+O2•−+2H+→2H2O2+O2 | Regulation of oxidative stress. Stress resistance or tolerance mechanisms |
Apoplast, cytosol, mitochondria, chloroplast, peroxisomes | |||
| Ascorbic Acid (AsA) | -Stress perception -Redox homeostasis -Regulation of oxidative stress -Improvement of plant stress tolerance |
Apoplast, cytosol mitochondria, chloroplast, vacuoles, peroxisomes, nucleus | [23,81][31][41] | [23,69][31][27] | ||
| Catalase (CAT) | H2O2→H2O+(1/2) O2 | Regulation of oxidative stress. Stress resistance or tolerance mechanisms. Plant metabolism |
Apoplast, cytosol, chloroplast, mitochondria, peroxisomes | |||
| Glutathione (GSH) | -Protect membranes. -Prevent protein oxidative denaturation under stress conditions -Substrate for glutathione peroxidase and gluthatione S-transferase -Metal chelator | [ | 23 | ,27,73][31 | Apoplast, cytosol, chloroplast, mitochondria, vacuole, peroxisome, nucleus][32][33] | |
| [ | 81 | ] | [41] | Ascorbate Peroxidase (APX) | H2O2+Asc→2H2O+DHA | Regulation of oxidative stress. Stress resistance or tolerance mechanisms |
| Proline (Pro) | -Osmoprotectant activity -Antioxidant capacity -Metal chelator -Signalling under abiotic and biotic stresses | Plant growth and physiology |
Apoplast, cytosol, mitochondria, peroxisomes, chloroplast | -Plant growth and development[74 | Apoplast, cytosol, mitochondria, chloroplast | [82,83][,75][34][35] |
| 42 | ] | [ | 43] | Monodehydroascorbate Reductase (MDHAR) | MDHA+NADPH→Asc+NADP+ | |
| Phenolic Compounds | -Antioxidant activity -Metal chelator -Protective and signalling functions against different stresses | Regulation of oxidative stress. Stress resistance or tolerance mechanisms |
Apoplast, cytosol, mitochondria, chloroplast | -Plant growth and development[ |
Ubiquitous | [84][44]23,76,77][31][36][37] |
| Dehydroascorbate Reductase (DHAR) | DHA+2GSH→Asc+GSSG | |||||
| Polyamines | -Antioxidant capacity -Plant growth and development. -Biotic and abiotic stress responses. | Regulation of oxidative stress Stress resistance or tolerance mechanisms Plant growth and development |
Apoplast, cytoplasm, mitochondria, chloroplast, peroxisomes | -Osmotic adjustment ability | Ubiquitous | [85,86][45][46][23,78,79][31][38][39] |
| Glutathione Reductase (GR) | GSSG+NADPH→2GSH+NADP+ | Regulation of oxidative stress. Stress resistance or tolerance mechanisms |
Apoplast, cytoplasm, mitochondria, chloroplast | [80][40] | ||
3. Apoplastic Proteins and Peptides in Modulating Plant–Pathogen Interactions. Microbe-Associated Molecular Patterns (MAMPs) of Proteic Nature
The most abundant proteins related to defense are the pathogen-related proteins (PR-proteins) that represent 23–33% of the total apoplastic fluid proteins (AFPs) [115][58]. The production and accumulation of PR-proteins are produced by various stresses, including biotic/biological stresses and abiotic/non-biological stresses [116][59]. The plant PR-proteins are divided into 17 groups (Table 3). Moreover, based on the amino acid sequence and the biochemical activity identified during the last decade, they also include novel peptide families. The PR peptides include proteinase inhibitors (PR-6 family), plant defensins (PR-12 family), thionins (PR-13 family), and lipid transfer proteins (PR-14 family) [117,118][60][61].| Families | Properties | References |
|---|---|---|
| PR-1 | Antifungal | [119][62] |
| PR-2 | β-1,3-glucanase | [120][63] |
| PR-3 | Chitinase type I, II, IV, V, VI, VII | [121][64] |
| PR-4 | Chitinase type I, II | [122][65] |
| PR-5 | Thaumatin- like | [39][66] |
| PR-6 | Proteinase- inhibitor | [123][67] |
| PR-7 | Endoproteinase | [124][68] |
| PR-8 | Chitinase type III | [125][69] |
| PR-9 | Peroxidase | [126][70] |
| PR-10 | Ribonuclease like | [127,128][71][72] |
| PR-11 | Chitinase, type I | [129][73] |
| PR-12 | Defensin | [130][74] |
| PR-13 | Thionin | [131][75] |
| PR-14 | Lipid- transfer protein | [132][76] |
| PR-15 | Oxalate oxidase | [133][77] |
| PR-16 | Oxalate oxidase-like | [123][67] |
| PR-17 | PRp27 Unknown | [134][78] |
3.1. Apoplastic Proteins Related to Plant Defense
3.2. Apoplastic Peptides Related to Plant Defense
3.3. Microbe-Associated Molecular Patterns of Proteic Nature
References
- Münch, E. Die Stoffbewegungen in der Pflanze; G. Fischer: Schaffhausen, Switzerland, 1930.
- Boyer, J.S. Cell wall biosynthesis and the molecular mechanism of plant enlargement. Funct. Plant Biol. 2009, 36, 383.
- Pauly, M.; Keegstra, K. Biosynthesis of the Plant Cell Wall Matrix Polysaccharide Xyloglucan. Annu. Rev. Plant Biol. 2016, 67, 235–259.
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 2018, 176, 2590–2600.
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335.
- Koch, W.; Kwart, M.; Laubner, M.; Heineke, D.; Stransky, H.; Frommer, W.B.; Tegeder, M. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 2003, 33, 211–220.
- McCully, M.E. Niches for bacterial endophytes in crop plants: A plant biologist’s view. Aust. J. Plant Physiol. 2001, 28, 983–990.
- Sattelmacher, B. The apoplast and its significance for plant mineral nutrition. New Phytol. 2001, 149, 167–192.
- Rico, A.; Preston, G.M. Pseudomonas syringae pv. tomato DC3000 Uses Constitutive and Apoplast-Induced Nutrient Assimilation Pathways to Catabolize Nutrients That Are Abundant in the Tomato Apoplast. Mol. Plant-Microbe Interact. 2008, 21, 269–282.
- Floerl, S.; Majcherczyk, A.; Possienke, M.; Feussner, K.; Tappe, H.; Gatz, C.; Feussner, I.; Kües, U.; Polle, A. Verticillium longisporum Infection Affects the Leaf Apoplastic Proteome, Metabolome, and Cell Wall Properties in Arabidopsis thaliana. PLoS ONE 2012, 7, e31435.
- Scalschi, L.; Llorens, E.; González-Hernández, A.I.; Valcárcel, M.; Gamir, J.; García-Agustín, P.; Vicedo, B.; Camañes, G. 1-Methyltryptophan Modifies Apoplast Content in Tomato Plants Improving Resistance Against Pseudomonas syringae. Front. Microbiol. 2018, 9, 2056.
- Hoson, T. Apoplast as the site of response to environmental signals. J. Plant Res. 1998, 111, 167–177.
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. 2010, 106, 185–197.
- Horst, W.J. The role of the apoplast in aluminium toxicity and resistance of higher plants: A review. Zeitschrift für Pflanzenernährung und Bodenkd. 1995, 158, 419–428.
- Geilfus, C.M. The pH of the Apoplast: Dynamic Factor with Functional Impact Under Stress. Mol. Plant 2017, 10, 1371–1386.
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006, 19, 827–837.
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95.
- Romero, F.M.; Rossi, F.R.; Gárriz, A.; Carrasco, P.; Ruíz, O.A. A Bacterial Endophyte from Apoplast Fluids Protects Canola Plants from Different Phytopathogens via Antibiosis and Induction of Host Resistance. Phytopathology 2019, 109, 375–383.
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 1–10.
- O’Leary, B.M.; Neale, H.C.; Geilfus, C.M.; Jackson, R.W.; Arnold, D.L.; Preston, G.M. Early changes in apoplast composition associated with defence and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae Pv. phaseolicola. Plant Cell Environ. 2016, 39.
- Doehlemann, G.; Hemetsberger, C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 2013, 198, 1001–1016.
- Yan, J.; Yu, H.; Li, B.; Fan, A.; Melkonian, J.; Wang, X.; Zhou, T.; Hua, J. Cell autonomous and non-autonomous functions of plant intracellular immune receptors in stomatal defense and apoplastic defense. PLoS Pathog. 2019, 15, e1008094.
- Caverzan, A.; Casassola, A.; Patussi Brammer, S. Reactive Oxygen Species and Antioxidant Enzymes Involved in Plant Tolerance to Stress. In Abiotic and Biotic Stress in Plants—Recent Advances and Future Perspectives; InTech: Nappanee, IN, USA, 2016.
- Kangasjärvi, S.; Kangasjärvi, J. Towards Understanding Extracellular ROS Sensory and Signaling Systems in Plants. Adv. Bot. 2014, 2014, 538946.
- Berwal, M.K.; Ram, C. Superoxide Dismutase: A Stable Biochemical Marker for Abiotic Stress Tolerance in Higher Plants. In Abiotic and Biotic Stress in Plants; IntechOpen: London, UK, 2019.
- Sgherri, C.; Quartacci, M.F.; Navari-Izzo, F. Early production of activated oxygen species in root apoplast of wheat following copper excess. J. Plant Physiol. 2007, 164, 1152–1160.
- García, G.; Clemente-Moreno, M.J.; Díaz-Vivancos, P.; García, M.; Hernández, J.A. The apoplastic and symplastic antioxidant system in onion: Response to long-term salt stress. Antioxidants 2020, 9, 67.
- Vanacker, H.; Harbinson, J.; Ruisch, J.; Carver, T.L.W.; Foyer, C.H. Antioxidant defences of the apoplast. Protoplasma 1998, 205, 129–140.
- Vanacker, H.; Carver, T.L.W.; Foyer, C.H. Pathogen-induced changes in the antioxidant status of the apoplast in barley leaves. Plant Physiol. 1998, 117, 1103–1114.
- Singh, B.N.; Singh, A.; Singh, S.P.; Singh, H.B. Trichoderma harzianum- mediated reprogramming of oxidative stress response in root apoplast of sunflower enhances defence against Rhizoctonia solani. Eur. J. Plant Pathol. 2011, 131, 121–134.
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53.
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12.
- Sharma, I.; Ahmad, P. Catalase: A Versatile Antioxidant in Plants. In Oxidative Damage to Plants: Antioxidant Networks and Signaling; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 131–148. ISBN 9780127999630.
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019.
- Sharova, E.I.; Medvedev, S.S.; Demidchik, V.V. Ascorbate in the Apoplast: Metabolism and Functions. Russ. J. Plant Physiol. 2020, 67, 207–220.
- Bindschedler, L.V.; Palmblad, M.; Cramer, R. Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry 2008, 69, 1962–1972.
- Chen, Z.; Gallie, D.R. Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 2006, 142, 775–787.
- Ding, H.; Wang, B.; Han, Y.; Li, S. The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J. Exp. Bot. 2020.
- Yousuf, P.Y.; Hakeem, K.U.R.; Chandna, R.; Ahmad, P. Role of Glutathione Reductase in Plant Abiotic Stress. In Abiotic Stress Responses in Plants; Springer: New York, NY, USA, 2012; pp. 149–158.
- Zechmann, B. Subcellular distribution of ascorbate in plants. Plant Signal. Behav. 2011, 6, 360–363.
- Hasanuzzaman, M.; Borhannuddin Bhuyan, M.H.M.; Anee, T.I.; Parvin, K.; Nahar, K.; Al Mahmud, J.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384.
- Saruhan Güler, N.; Sağlam, A.; Demiralay, M.; Kadioğlu, A.; Saruhan Güler, N.; Sağlam, A.; Demiralay, M.; Kadioğlu, A. Apoplastic and symplastic solute concentrations contribute to osmotic adjustment in bean genotypes during drought stress. Turk J Biol 2012, 36, 151–160.
- Dar, M.I.; Naikoo, M.I.; Rehman, F.; Naushin, F.; Khan, F.A. Proline accumulation in plants: Roles in stress tolerance and plant development. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Springer: Delhi, India, 2015; pp. 155–166. ISBN 9788132226161.
- Babenko, L.M.; Smirnov, O.E.; Romanenko, K.O.; Trunova, O.K.; Kosakivska, I.V. Phenolic compounds in plants: Biogenesis and functions. Ukr. Biochem. J. 2019, 91, 5–18.
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945.
- Takahashi, T.; Kakehi, J.-I. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 2009, 105, 1–6.
- Podgórska, A.; Burian, M.; Szal, B. Extra-cellular but extra-ordinarily important for cells: Apoplastic reactive oxygen species metabolism. Front. Plant Sci. 2017, 8.
- Salguero, J.; Böttger, M. Secreted catalase activity from roots of developing maize (Zea mays L.) seedlings. Protoplasma 1995, 184, 72–78.
- Parra-Lobato, M.C.; Fernandez-Garcia, N.; Olmos, E.; Alvarez-Tinaut, M.C.; Gómez-Jiménez, M.C. Methyl jasmonate-induced antioxidant defence in root apoplast from sunflower seedlings. Environ. Exp. Bot. 2009, 66, 9–17.
- Zhou, Y.; Liu, S.; Yang, Z.; Yang, Y.; Jiang, L.; Hu, L. CsCAT3, a catalase gene from Cucumis sativus, confers resistance to a variety of stresses to Escherichia coli. Biotechnol. Biotechnol. Equip. 2017, 31, 886–896.
- Fernández-Crespo, E.; Gómez-Pastor, R.; Scalschi, L.; Llorens, E.; Camañes, G.; García-Agustín, P. NH4 + induces antioxidant cellular machinery and provides resistance to salt stress in citrus plants. Trees 2014, 28, 1693–1704.
- De Pinto, M.C.; De Gara, L. Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J. Exp. Bot. 2004, 55, 2559–2569.
- Asada, K. Ascorbate peroxidase—A hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 1992, 85, 235–241.
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017, 8.
- O’Brien, J.A.; Daudi, A.; Finch, P.; Butt, V.S.; Whitelegge, J.P.; Souda, P.; Ausubel, F.M.; Bolwell, G.P. A Peroxidase-Dependent Apoplastic Oxidative Burst in Cultured Arabidopsis Cells Functions in MAMP-Elicited Defense. Plant Physiol. 2012, 158, 2013–2027.
- Saruhan, N.; Terzi, R.; Saǧlam, A.; Kadioǧlu, A. Scavenging of reactive oxygen species in apoplastic and symplastic areas of rolled leaves in Ctenanthe setosa under drought stress. Acta Biol. Hung. 2010, 61, 282–298.
- Diaz-Vivancos, P.; Rubio, M.; Mesonero, V.; Periago, P.M.; Barceló, A.R.; Martínez-Gómez, P.; Hernández, J.A. The apoplastic antioxidant system in Prunus: Response to long-term plum pox virus infection. J. Exp. Bot. 2006, 57, 3813–3824.
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; do Céu Silva, M.; Cardoso, F.M.H.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6, 478.
- Tuzun, S.; Somanchi, A.; der Hoorn, R. The Possible Role of PR proteins in Multigenic and Induced Systemic Resistance. In Multigenic and Induced Systemic Resistance in Plants; Springer: Berlin, Germany, 2008; Volume 59 SRC-, pp. 112–142.
- Dempsey, D.A.; Silva, H.; Klessig, D.F. Engineering disease and pest resistance in plants. Trends Microbiol. 1998, 6, 54–61.
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950.
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879.
- Hong, T.; Meng, M. Biochemical characterization and antifungal activity of an endo-3-beta-glucanase of Paenibacillus sp. Isol. from Gard. soil. Appl. Microbiol. Biotechnol. 2003, 61, 472–478.
- Yang, Q.; Gong, Z. Purification and characterization of an ethylene-induced antifungal protein from leaves of guilder rose (Hydrangea macrophylla). Protein Expr. Purif. 2002, 24, 76–82.
- Yang, Y.-X.; Ahammed, G.; Wu, C.; Fan, S.; Zhou, Y.-H. Crosstalk among Jasmonate, Salicylate and Ethylene Signaling Pathways in Plant Disease and Immune Responses. Curr. Protein Pept. Sci. 2015, 16, 450–461.
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162.
- Naz, R.; Bano, A.; Wilson, N.L.; Guest, D.; Roberts, T.H. Pathogenesis-related protein expression in the apoplast of wheat leaves protected against leaf rust following application of plant extracts. Phytopathology 2014, 104, 933–944.
- Tornero, P.; Conejero, V.; Vera, P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J. Biol. Chem. 1997, 272, 14412–14419.
- Simmons, C. The Physiology and Molecular Biology of Plant 1,3-β-D-Glucanases and 1,3;1,4-β-D-Glucanases. Crit. Rev. Plant Sci. 1994, 13, 325–387.
- Van Loon, L.C. Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 1997, 103, 753–765.
- Sugawara, T.; Trifonova, E.A.; Kochetov, A.V.; Kanayama, Y. Expression of an extracellular ribonuclease gene increases resistance to Cucumber mosaic virus in tobacco. BMC Plant Biol. 2016, 16.
- Filipenko, E.; Kochetov, A.; Kanayama, Y.; Malinovsky, V.; Shumny, V. PR-proteins with ribonuclease activity and plant resistance against pathogenic fungi. Russ. J. Genet. Appl. Res. 2013, 10 SRC-, 474–480.
- Bravo, J.M.; Campo, S.; Murillo, I.; Coca, M.; San Segundo, B. Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 2003, 52, 745–759.
- Penninckx, I.A.; Eggermont, K.; Terras, F.R.; Thomma, B.P.; De Samblanx, G.W.; Buchala, A.; Métraux, J.P.; Manners, J.M.; Broekaert, W.F. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996, 8, 2309–2323.
- Stec, B. Plant thionins--the structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385.
- Kido, E.A.; Pandolfi, V.; Houllou-Kido, L.M.; Andrade, P.P.; Marcelino, F.C.; Nepomuceno, A.L.; Abdelnoor, R.V.; Burnquist, W.L.; Benko-Iseppon, A.M. Plant Antimicrobial Peptides: An Overview of SuperSAGE Transcriptional Profile and a Functional Review. Curr. Protein Pept. Sci. 2010, 11, 220–230.
- Hu, X.; Bidney, D.L.; Yalpani, N.; Duvick, J.P.; Crasta, O.; Folkerts, O.; Lu, G. Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol. 2003, 133, 170–181.
- Christensen, A.B.; Cho, B.H.O.; Næsby, M.; Gregersen, P.L.; Brandt, J.; Madriz-Ordeñana, K.; Collinge, D.B.; Thordal-Christensen, H. The molecular characterization of two barley proteins establishes the novel PR-17 family of pathogenesis-related proteins. Mol. Plant Pathol. 2002, 3, 135–144.
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97.
- Liu, Q.; Xue, Q. Computational identification of novel PR-1-type genes in Oryza sativa. J. Genet. 2006, 85, 193–198.
- Payne, G.; Ward, E.; Gaffney, T.; Goy, P.A.; Moyer, M.; Harper, A.; Meins, F.; Ryals, J. Evidence for a third structural class of β-1,3-glucanase in tobacco. Plant Mol. Biol. 1990, 15, 797–808.
- Leubner-Metzger, G.; Meins, F. 3. Functions and Regulation of Plant ß-1,3-glucanases (PR-2); CRC Press LLC: Boca Raton, FL, USA, 1999.
- Dyakov, Y.T.; Dzhavakhiya, V.G.; Korpela, T. Comprehensive and Molecular Phytopathology; Elsevier: Amsterdam, The Netherlands, 2007; ISBN 9780080469331.
- Castresana, C.; de Carvalho, F.; Gheysen, G.; Habets, M.; Inzé, D.; Van Montagu, M. Tissue-specific and pathogen-induced regulation of a Nicotiana plumbaginifolia beta-1,3-glucanase gene. Plant Cell 1990, 2, 1131–1143.
- Alonso, E.; de Carvalho Niebel, F.; Obregon, P.; Gheysen, G.; Inze, D.; Van Montagu, M.; Castresana, C. Differential in vitro DNA binding activity to a promoter element of the gn1 beta-1,3-glucanase gene in hypersensitively reacting tobacco plants. Plant J. 1995, 7, 309–320.
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990.
- Vázquez-Garcidueñas, S.; Leal-Morales, C.A.; Herrera-Estrella, A. Analysis of the beta-1,3-Glucanolytic System of the Biocontrol Agent Trichoderma harzianum. Appl. Environ. Microbiol. 1998, 64, 1442–1446.
- Ait-Lahsen, H.; Soler, A.; Rey, M.; De La Cruz, J.; Monte, E.; Llobell, A. An Antifungal Exo-α-1,3-Glucanase (AGN13.1) from the Biocontrol Fungus Trichoderma harzianum. Appl. Environ. Microbiol. 2001, 67, 5833–5839.
- Kasprzewska, A. Plant chitinases--regulation and function. Cell. Mol. Biol. Lett. 2003, 8, 809–824.
- Saikia, R.; Singh, B.P.; Kumar, R.; Arora, D.K. Detection of pathogenesis-related proteins-chitinase and β-1,3-glucanase in induced chickpea. Curr. Sci. 2005, 84, 659–663.
- Garg, N.; Gupta, H. Isolation and purification of fungal pathogen (Macrophomina phaseolina) induced chitinase from moth beans (Phaseolus aconitifolius). J. Pharm. Bio Allied Sci. 2010, 2, 38.
- Fukamizo, T.; Chie, S.; Masahiro, T. Plant chitinases: Structure-function relationships and their physiology. Foods Food Ingredients J. 2003, 208, 631–632.
- Gooday, G.W. Aggressive and defensive roles for chitinases. EXS 1999, 87, 157–169.
- Afroz, A.; Ali, G.M.; Mir, A.; Komatsu, S. Application of proteomics to investigate stress-induced proteins for improvement in crop protection. Plant Cell Rep. 2011, 30, 745–763.
- Cantu, D.; Vicente, A.R.; Labavitch, J.M.; Bennett, A.B.; Powell, A.L.T. Strangers in the matrix: Plant cell walls and pathogen susceptibility. Trends Plant Sci. 2008, 13, 610–617.
- Petriccione, M.; Salzano, A.M.; Di Cecco, I.; Scaloni, A.; Scortichini, M. Proteomic analysis of the Actinidia deliciosa leaf apoplast during biotrophic colonization by Pseudomonas syringae pv. actinidiae. J. Proteom. 2014, 101, 43–62.
- Pechanova, O.; Hsu, C.-Y.; Adams, J.P.; Pechan, T.; Vandervelde, L.; Drnevich, J.; Jawdy, S.; Adeli, A.; Suttle, J.C.; Lawrence, A.M.; et al. Apoplast proteome reveals that extracellular matrix contributes to multistress response in poplar. BMC Genom. 2010, 11, 674.
- Delaunois, B.; Colby, T.; Belloy, N.; Conreux, A.; Harzen, A.; Baillieul, F.; Clément, C.; Schmidt, J.; Jeandet, P.; Cordelier, S. Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biol. 2013, 13, 24.
- Floerl, S.; Druebert, C.; Majcherczyk, A.; Karlovsky, P.; Kües, U.; Polle, A. Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol. 2008, 8, 129.
- Segarra, C.I.; Casalongué, C.A.; Pinedo, M.L.; Ronchi, V.P.; Conde, R.D. A germin-like protein of wheat leaf apoplast inhibits serine proteases. J. Exp. Bot. 2003, 54, 1335–1341.
- Joosten, M.H.A.J.; De Wit, P.J.G.M. Identification of Several Pathogenesis-Related Proteins in Tomato Leaves Inoculated with Cladosporium fulvum (syn. Fulvia fulva ) as 1,3-β-Glucanases and Chitinases. Plant Physiol. 1989, 89, 945–951.
- Han, L.B.; Li, Y.B.; Wang, F.X.; Wang, W.Y.; Liu, J.; Wu, J.H.; Zhong, N.Q.; Wu, S.J.; Jiao, G.L.; Wang, H.Y.; et al. The Cotton Apoplastic Protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen verticillium dahliae. Plant Cell 2019, 31, 520–536.
- Hon, W.-C.; Griffith, M.; Mlynarz, A.; Kwok, Y.C.; Yang, D.S. Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol. 1995, 109, 879–889.
- Rajam, M.V.; Chandola, N.; Goud, P.S.; Singh, D.; Kashyap, V.; Choudhary, M.L.; Sihachakr, D. Thaumatin gene confers resistance to fungal pathogens as well as tolerance to abiotic stresses in transgenic tobacco plants. Biol. Plant. 2007, 51, 135–141.
- Islam, M.A.; Sturrock, R.N.; Holmes, T.A.; Ekramoddoullah, A.K.M. Ultrastructural studies of Phellinus sulphurascens infection of Douglas-fir roots and immunolocalization of host pathogenesis-related proteins. Mycol. Res. 2009, 113, 700–712.
- Wang, X.; Tang, C.; Deng, L.; Cai, G.; Liu, X.; Liu, B.; Han, Q.; Buchenauer, H.; Wei, G.; Han, D.; et al. Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust fungus. Physiol. Plant. 2010, 139, 27–38.
- Shatters, R.G.; Boykin, L.M.; Lapointe, S.L.; Hunter, W.B.; Weathersbee, A.A. Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. J. Mol. Evol. 2006, 63, 12–29.
- Dunwell, J.M.; Khuri, S.; Gane, P.J. Microbial Relatives of the Seed Storage Proteins of Higher Plants: Conservation of Structure and Diversification of Function during Evolution of the Cupin Superfamily. Microbiol. Mol. Biol. Rev. 2000, 64, 153–179.
- Godfrey, D.; Able, A.J.; Dry, I.B. Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: A possible role in defense? Mol. Plant. Microbe. Interact. 2007, 20, 1112–1125.
- Regente, M.; Pinedo, M.; San Clemente, H.; Balliau, T.; Jamet, E.; de la Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 2017, 68, 5485–5495.
- Wojtaszek, P.; Stobiecki, M.; Bolwell, G.P. Changes in the composition of exocellular proteins of suspension-cultured Lupinus albus cells in response to fungal elicitors or CuCl2. J. Exp. Bot. 1997, 48, 2015–2021.
- Dani, V.; Simon, W.J.; Duranti, M.; Croy, R.R.D.R.D. Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 2005, 5, 737–745.
- Jaswanthi, N.; Krishna, M.S.R.; Sahitya, U.L.; Suneetha, P. Apoplast proteomic analysis reveals drought stress-responsive protein datasets in chilli (Capsicum annuum L.). Data Br. 2019, 25, 104041.
- Dunwell, J.M.; Gibbings, J.G.; Mahmood, T.; Saqlan Naqvi, S.M. Germin and germin-like proteins: Evolution, structure, and function. CRC. Crit. Rev. Plant Sci. 2008, 27, 342–375.
- Somssich, I.; Hahlbrock, K. Pathogen defence in plants — a paradigm of biological complexity. Trends Plant Sci. 1998, 3, 86–90.
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116.
- Khuri, S.; Bakker, F.T.; Dunwell, J.M. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 2001, 18, 593–605.
- Wang, T.; Chen, X.; Zhu, F.; Li, H.; Li, L.; Yang, Q.; Chi, X.; Yu, S.; Liang, X. Characterization of Peanut Germin-Like Proteins, AhGLPs in Plant Development and Defense. PLoS ONE 2013, 8, e61722.
- Amini, F.; Ehsanpour, A.A.; Hoang, Q.T.; Shin, J.S. Protein pattern changes in tomato under in vitro salt stress. Russ. J. Plant Physiol. 2007, 54, 464–471.
- Van der Hoorn, R.A.L. Plant Proteases: From Phenotypes to Molecular Mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223.
- Tian, M.; Win, J.; Song, J.; Van Der Hoorn, R.; van der Knaap, E.; Kamoun, S. A Phytophthora infestans Cystatin-Like Protein Targets a Novel Tomato Papain-Like Apoplastic Protease. Plant Physiol. 2006, 143, 364–377.
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. Subtilisin-like proteases in plant–pathogen recognition and immune priming: A perspective. Front. Plant Sci. 2014, 5, 739.
- Tian, M.; Benedetti, B.; Kamoun, S. A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005, 138, 1785–1793.
- Ramírez, V.; López, A.; Mauch-Mani, B.; Gil, M.J.; Vera, P. An Extracellular Subtilase Switch for Immune Priming in Arabidopsis. PLoS Pathog. 2013, 9, e1003445.
- Terras, F.R.G.; Schoofs, H.M.E.; Thevissen, K.; Osborn, R.W.; Vanderleyden, J.; Cammue, B.P.A.; Broekaert, W.F. Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2S albumins and by barley trypsin inhibitors. Plant Physiol. 1993, 103, 1311–1319.
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406.
- Planas-Marquès, M.; Bernardo-Faura, M.; Paulus, J.; Kaschani, F.; Kaiser, M.; Valls, M.; Van Der Hoorn, R.A.L.; Coll, N.S. Protease Activities Triggered by Ralstonia solanacearum Infection in Susceptible and Tolerant Tomato Lines. Mol. Cell. Proteom. 2018, 17, 1112–1125.
- Ziemann, S.; van der Linde, K.; Lahrmann, U.; Acar, B.; Kaschani, F.; Colby, T.; Kaiser, M.; Ding, Y.; Schmelz, E.; Huffaker, A.; et al. An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 2018, 4, 172–180.
- Schulze Hüynck, J.; Kaschani, F.; van der Linde, K.; Ziemann, S.; Müller, A.N.; Colby, T.; Kaiser, M.; Misas Villamil, J.C.; Doehlemann, G. Proteases underground: Analysis of the maize root apoplast identifies organ specific papain-like cysteine protease activity. Front. Plant Sci. 2019, 10, 473.
- Krüger, J.; Thomas, C.M.; Golstein, C.; Dixon, M.S.; Smoker, M.; Tang, S.; Mulder, L.; Jones, J.D.G. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 2002, 296, 744–747.
- Lozano-Torres, J.L.; Wilbers, R.H.P.; Gawronski, P.; Boshoven, J.C.; Finkers-Tomczak, A.; Cordewener, J.H.G.; America, A.H.P.; Overmars, H.A.; Van ’t Klooster, J.W.; Baranowski, L.; et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA 2012, 109, 10119–10124.
- Lay, F.; Anderson, M. Defensins - Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101.
- Bohlmann, H. The role of thionins in plant protection. CRC. Crit. Rev. Plant Sci. 1994, 13, 1–16.
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Osborn, R.W. Plant defensins: Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995, 108, 1353–1358.
- Kaur, J.; Fellers, J.; Adholeya, A.; Velivelli, S.L.S.; El-Mounadi, K.; Nersesian, N.; Clemente, T.; Shah, D. Expression of apoplast-targeted plant defensin MtDef4.2 confers resistance to leaf rust pathogen Puccinia triticina but does not affect mycorrhizal symbiosis in transgenic wheat. Transgenic Res. 2017, 26, 37–49.
- Hsiao, P.Y.; Cheng, C.P.; Koh, K.W.; Chan, M.T. The Arabidopsis defensin gene, AtPDF1.1, mediates defence against Pectobacterium carotovorum subsp. carotovorum via an iron-withholding defence system. Sci. Rep. 2017, 7.
- Koike, M.; Okamoto, T.; Tsuda, S.; Imai, R. A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem. Biophys. Res. Commun. 2002, 298, 46–53.
- Boutrot, F.; Chantret, N.; Gautier, M.F. Genome-wide analysis of the rice and arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 2008, 9.
- Kader, J.-C. LIPID-TRANSFER PROTEINS IN PLANTS. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 627–654.
- De O. Carvalho, A.; de S. Teodoro, C.E.; Da Cunha, M.; Okorokova-Facanha, A.L.; Okorokov, L.A.; Fernandes, K.V.S.; Gomes, V.M. Intracellular localization of a lipid transfer protein in Vigna unguiculata seeds. Physiol. Plant. 2004, 122, 328–336.
- Kusumawati, L.; Imin, N.; Djordjevic, M.A. Characterization of the secretome of suspension cultures of medicago species reveals proteins important for defense and development. J. Proteome Res. 2008, 7, 4508–4520.
- Pagnussat, L.; Burbach, C.; Baluska, F.; de la Canal, L. An extracellular lipid transfer protein is relocalized intracellularly during seed germination. J. Exp. Bot. 2012, 63, 6555–6563.
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Lipid Transfer Proteins As Components of the Plant Innate Immune System: Structure, Functions, and Applications. Acta Naturae 2016, 8, 47–61.
- Sakurai, N. Dynamic function and regulation of apoplast in the plant body. J. Plant Res. 1998, 111, 133–148.
- Lee, S.B.; Go, Y.S.; Bae, H.J.; Park, J.H.; Cho, S.H.; Cho, H.J.; Lee, D.S.; Park, O.K.; Hwang, I.; Suh, M.C. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen alternaria brassicicola. Plant Physiol. 2009, 150, 42–54.
- Sarowar, S.; Kim, Y.J.; Kim, K.D.; Hwang, B.K.; Ok, S.H.; Shin, J.S. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 2009, 28, 419–427.
- Maldonado, A.M.; Doerner, P.; Dixonk, R.A.; Lamb, C.J.; Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 2002, 419, 399–403.
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983.
- Nürnberger, T.; Brunner, F.; Kemmerling, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198.
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276.
- Gómez-Gómez, L.; Felix, G.; Boller, T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999, 18, 277–284.
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR Receptor–like Kinase Involved in the Perception of the Bacterial Elicitor Flagellin in Arabidopsis. Mol. Cell 2000, 2, 1003–1011.
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628.
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.G.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767.
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507.
- Mott, G.A.; Middleton, M.A.; Desveaux, D.; Guttman, D.S. Peptides and small molecules of the plant-pathogen apoplastic arena. Front. Plant Sci. 2014, 5, 677.
- Wang, L.; Albert, M.; Einig, E.; Fürst, U.; Krust, D.; Felix, G. The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2016, 2.
- Felix, G.; Grosskopf, D.G.; Regenass, M.; Basse, C.W.; Boller, T. Elicitor-induced ethylene biosynthesis in tomato cells: Characterization and use as a bioassay for elicitor action. Plant Physiol. 1991, 97, 19–25.
- Yano, A.; Suzuki, K.; Uchimiya, H.; Shinshi, H. Induction of hypersensitive cell death by a fungal protein in cultures of tobacco cells. Mol. Plant-Microbe Interact. 1998, 11, 115–123.
- Enkerli, J.; Felix, G.; Boller, T. The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 1999, 121, 391–397.
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615.
- Ranf, S.; Gisch, N.; Schäffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Sánchez-Carballo, P.M.; Zähringer, U.; Hückelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433.
- Willmann, R.; Lajunen, H.M.; Erbs, G.; Newman, M.-A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.-J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829.
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502.
