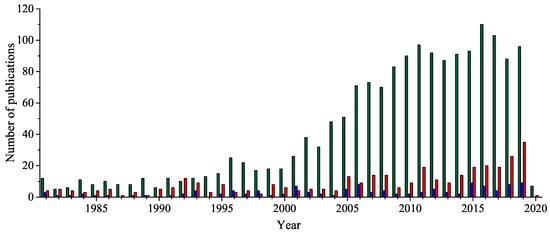
Five-membered 1,2,4-oxadiazole heterocyclic ring has received considerable attention because of its unique bioisosteric properties and an unusually wide spectrum of biological activities. Thus, it is a perfect framework for the novel drug development. After a century since the 1,2,4-oxadiazole have been discovered, the uncommon potential attracted medicinal chemists’ attention, leading to the discovery of a few presently accessible drugs containing 1,2,4-oxadiazole unit. It is worth noting that the interest in a 1,2,4-oxadiazoles’ biological application has been doubled in the last fifteen years. Herein, after a concise historical introduction, we present a comprehensive overview of the recent achievements in the synthesis of 1,2,4-oxadiazole-based compounds and the major advances in their biological applications in the period of the last five years as well as brief remarks on prospects for further development.
- 1.2.4-oxadiazole
- synthetic methods
- drug design
- drug discovery
- structure-activity relationship
- medicinal application
1. Introduction
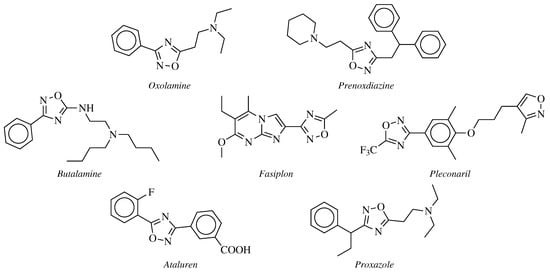
2. Historical Remarks—1,2,4-Oxadiazole
3. Anticancer Agents
Every year cancer impacts about 20 million people all over the world resulting in deaths counting in millions (Figure 5). Unfortunately, a number of new cancer cases is still rising and almost 30 million people will be diagnosed with carcinoma by 2040 in high-developed countries [50][30]. For that reason, finding new cancer treatments or effective drugs is one of the greatest needs of the current community and a challenge for modern medicine. Biological evaluation of 1,2,4-oxadiazoles revealed that some of their derivatives are potent anticancer agents. The greatest breakthrough came with the discovery of 3,5-diarylsubstituted derivatives of 1,2,4-oxadiazole as a new series of apoptosis inducers [51][31]. Since then, exploration of the anticancer activity of 1,2,4-oxadiazole derivatives has been started resulting in a creation of a wide library of compounds [52,53][32][33].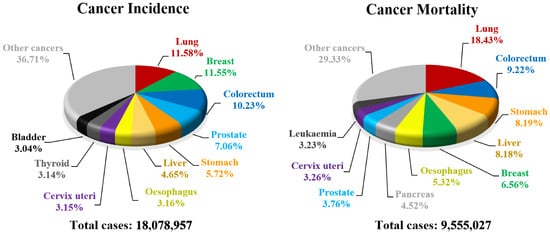
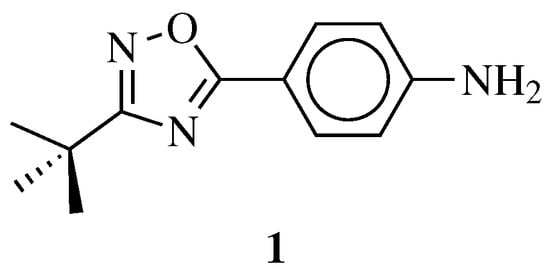
| General Structure | Substituents | The Most Active Derivatives | Activity | Ref. |
 |
R1 = H, NH2 and other (see Ref.); R2 = H or phenyl. |
 |
IC50 values of 2.76 and 9.27 μM against OVXF 899 and PXF 1752 cancer cell lines, respectively. | [35] |
| General Structure | Substituents | The Most Active Derivatives | Activity | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
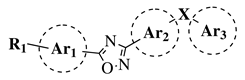 |
R1 = H, OH, OCH3, NH2, NHAc, NH3Cl, NHMs, NH-nBu, NH-tBu, NHCOPh, NH-iPr, PO3H2, PO(OEt)2, SO2NH2, CONH2, COOH, COOCH3 F, Cl, Br, I, NO2, ethynyl or CN; Ar1 = phenyl, benzyl, 2-pyrole, 3-pyridyl, 4-pyridyl, 5-indole, 3-pyrrazole, 2-imidazole and many others (see Ref.); Ar2, Ar3 = p-phenylene, 6-indole, 2-pyridyl, 6-chromene, carbazole, N-phenylpiperazine, N-phenylmorpholine and many others (see Ref.); X = NH, CH2, O, CO, NBn, SO or SO2. |
 |
MIC50 values <4 μg/mL against over 210 diverse, MRSA and VRE strains. | [96,98,99,103][65][67][68][72] | |||||
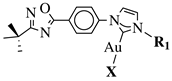 |
X = Cl or Br; R1 = methyl, benzyl, 2-pyridinyl or anthracen-9-ylmethyl. |
 |
IC50 values of 3 nM against LXFA 629 and MAXF 401 cancer cell lines, respectively. | ||||||
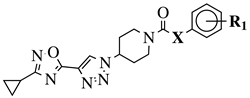 |
X = NH or none; R1 = H, 3-chloro-4-fluorophenyl, 2-chlorophenyl, 2-ethyl, 4-ethyl, 5-bromo-2-fluorophenyl or 2-methylpyridin-5-yl. |
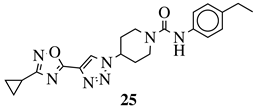 |
Grown inhibition zone within 20–25 mm against S. aureus, B. subtilis, E. coli, P. vulgaris, P | [36] | |||||
| . | aeruginosa | , | C. albicans. | [104][73] | 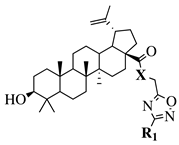 |
X = O or NH; R1 = phenyl, benzyl, 2-chlorophenyl, 4-fluorophenyl, 2-mehylphenyl, 4-bromophenyl, 4-methylphenyl, 4-methoxyphenyl, 4-pyridinyl, 2-methoxyphenyl, 2-benzyloxyphenyl or 3-pyridinyl. |
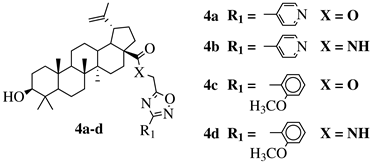 |
||
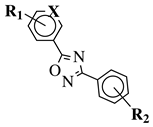 |
R1 = H, 2-chloro or 3-chloro; X = CH or N; R2 = H, 2-nitro, 2-chloro, 3-bromo, 2-chloro-5-nitro, 2-bromo, 3-nitro, 2-iodo, 3,5-dinitro, 4-nitro or 2-hydroxy. | IC | 50 values between 26.1–34.3 μM against Colo 205, Hep G2 and Hela cell lines. | 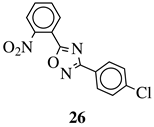 |
MIC value of 60 μM against E. [37][38] | ||||
| coli | . | [ | 105 | ][74] | 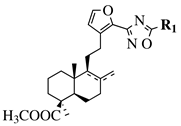 |
R1 = methyl, chloromethyl or phenyl. | |||
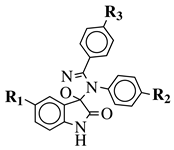 |  |
R1 = H, F, Cl, Br, I, methyl, ethyl, methoxy or iPr; R2 = H, methyl, methoxy, iPr, F, Cl, Br or I; R3 = H, F, Cl, Br, nitro, iPr, OBn, methoxy, ethoxy or CN.GI |
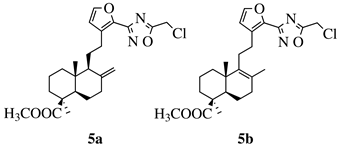
 50 values of 0.08 ( 50 values of 0.08 ( |
MIC value of 64 μ5a) and 0.34 (5b) μM against CEM-13 cell line. | g/mL against S[39] | ||||
| . | epidermidis | . | [ | 106][75] | 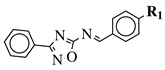 |
R1 = H, Br, Cl, F, methoxy or NH2. | |||
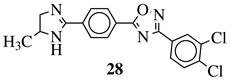
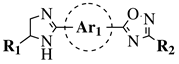 |
R1 = H or methyl; | 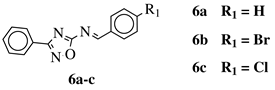 |
Ar1 = p-phenylene or m-phenylene; R2 = methyl, cyclopropyl, 2-thienyl, 2-chlorophenyl, 3-chlorophenyl, 3,4-dichlorophenyl, 4-ethylphenyl, 4-t-butylphenyl, 4-methylphenyl, 3,4,-dimethylphenyl and many others (see Ref.).CC50 values of 137.3, 79.0 and 140.3 μM against Ca9-22 cell line, respectively. |
 |
MIC values in a range 8–16 μg/mL toward S. aureus, B. subtilis, E. coli, P. fluorescent.[40] | ||||
| [ | 107 | ] | [ | 76] |  |
R1 = H, 2-chloro, 3-chloro, 4-chloro, 4-nitro, 4-methyl, 4-methoxy, 4-trifluoromethyl, 2-bromo, 3-bromo, 4-bromo or 4-fluoro; R2 = N(CH3)2, N(C2H5)2, pyrrolidine-1-yl, azepan-1-yl, morpholin-1-yl, thiomorpholine-1-yl, N-methylpiperazin-1-yl, N-phenylpiperazin-1-yl, 3-bromopropan-1-yl or 3-chloropropan-1-yl. |
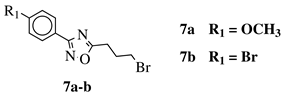 |
80% of death of NB4, K562 and MDA-MB-231 cancer cell lines at 25 (7a) and 10 (7b) μM. | [41] |
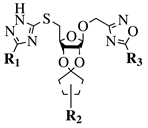 |
R1 = H or NH2; R2 = isopropylidene or cyclopentylidene; R3 = 4-nitrophenyl, 4-chlorophenyl or 3,4,5-trimethylphenyl. |
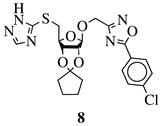 |
GI50 of 4.5 μM against WiDr cancer cell line. | [42] | |||||
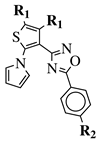 |
R1 = CH3 or —(CH2)4—; R2 = H, Cl, Br, methyl or methoxy. |
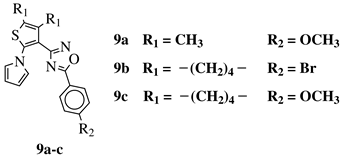 |
IC50 values of 0.48 (9a), 0.78 (9b), 0.19 (9c) μM against MCF-7 cancer cell line. | [43] | |||||
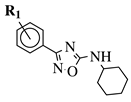 |
R1 = H, 3-methyl, 4-methyl, 3-bromo, 4-methoxy, 4-trifluoromethyl, 4-chloro, 4-bromo or 4-fluoro. | 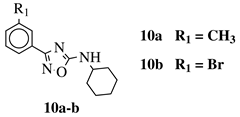 |
IC50 values between 13.6–48.37 μM against HCT-116, PC-3, SNB-19, B16F10, L929 cell lines. | [44] | |||||
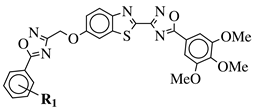 |
R1 = H, 3,4,5-trimethoxy, 4-methoxy, 4-chloro, 4-bromo, 4-fluoro, 4-trifluoromethyl, 4-nitro, 4-cyano or 4-methyl. | 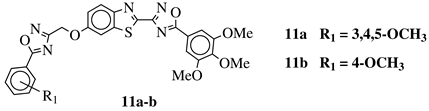 |
IC50 values in a range from 0.11 to 2.09 μM against MCF-7, A375 and HT-29 cancer cell lines. | [45] | |||||
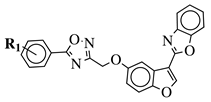 |
R1 = H, 3,4,5-trimethoxy, 4-methoxy, 4-chloro, 4-bromo, 4-fluoro, 4-trifluoromethyl, 4-nitro, 4-cyano or 4-methyl. | 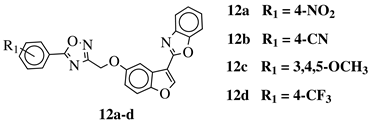 |
IC50 values between 0.011–1.89 μM against A549, MCF-7, A375 and HT-29 cancer cell lines. | [46] | |||||
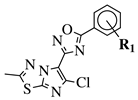 |
R1 = H, 3,4,5-trimethoxy, 4-methoxy, 4-chloro, 4-bromo, 4-fluoro, 4-trifluoromethyl, 4-nitro, 3-nitro or 4-methyl. | 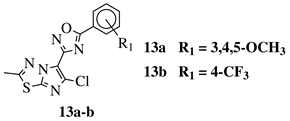 |
IC50 values in a range of 0.11–1.47 μM against A375, MCF-7 and ACHN cancer cell lines. | [47] | |||||
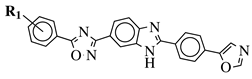 |
R1 = H, 3,4,5-trimethoxy, 4-methoxy, 4-chloro, 4-bromo, 4-fluoro, 4-trifluoromethyl, 4-nitro, 4-cyano or 4-methyl. | 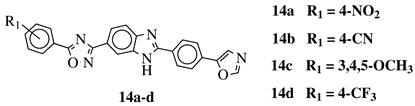 |
IC50 values between 0.12–2.78 μM against MCF-7, A549 and A375 cancer cell lines. | [48] | |||||
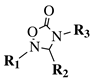 |
R1 = methyl, phenyl, 4-fluorophenyl, benzyl or 4-methoxbenzyl; R2 = phenyl, 9-phenanthryl or 4-pyridinyl; R3 = 4-nitrophenyl, 4-chlorophenyl, 4-trifluoromethylphenyl or 4-fluorophenyl. |
 |
IC50 value of 10.38 μM toward MCF-7 cancer cell line. | [49] | |||||
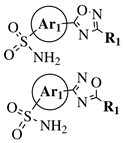 |
R1 = methyl, phenyl, 2-methoxyphenyl, 3-methoxyphenyl, 4-t-butylphenyl, 4-methylphenyl, 2-methoxyphenyl, 3-methoxyphenyl, cyclopropyl, 2-pyridinyl, 3-pyridinyl, 4-pyridinyl, 2-thienyl, 3-thienyl, 4-cyanophenyl, 2-fluorophenyl, 3-fluorophenyl, 4-fluorophenyl, 2-chlorophenyl or 3,4-dichlorophenyl; Ar1 = p-phenylene, m-phenylene, p-methoxyphenylene or 2,4-thienyl. |
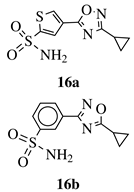 |
Ki value of 89 pm and 0.75 nm (hCA IX and hCA II, respectively) for 16a in CO2 hydration stopped-flow biochemical assay. 16b showed high selectivity toward PANC-1 cancer cell line. |
[50][51] | |||||
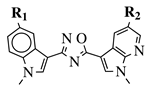 |
R1 = H, F, Cl, Br or methoxy; R2 = H, F or Br. |
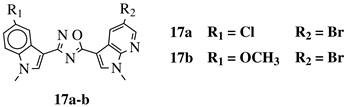 |
IC50 values of 0.65 (17a) and 2.41 μM (17b) against MCF-7 cancer cell line. | [52] | |||||
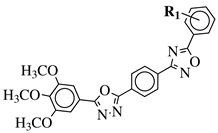 |
R1 = H, 3,4,5-trimethoxy, 4-methoxy, 4-chloro, 4-bromo, 4-fluoro, 4-nitro, 3-nitro, 4-cyano or 4-trifluoromethyl. | 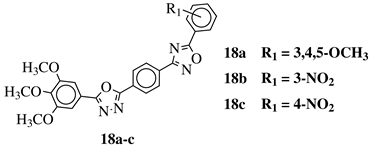 |
IC50 values in a range of 0.45–2.11 μM against MCF-7, A549, MDA-MB-231 cancer cell lines. | [53] | |||||
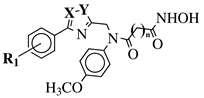 |
X, Y = N, O or O, N; n = 5 or 6; R1 = H, 2-methyl, 4-methyl, 4-methoxy, 2-fluoro, 3-fluoro, 4-fluoro, 4-bromo or 4-nitro. |
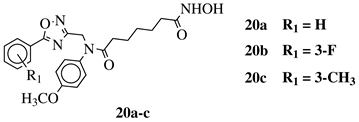 |
IC50 values of 8.2, 10.5, 12.1 nM (20a, 20b, 20c, respectively) toward HDAC-1. | [54][55] | |||||
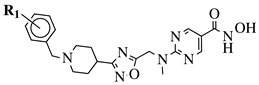 |
R1 = H, 4-methyl, 3-methyl, 2-fluoro, 4-fluoro, 2,4-difluoro, 2-chloro, 4-cyano, 4-trifluoromethyl or 2-chloro-4-fluoro. | 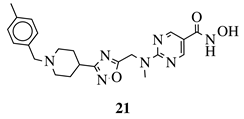 |
IC50 values of 1.8, 3.6 and 3.0 nM against HDAC-1, -2 and -3, respectively. | [56] | |||||
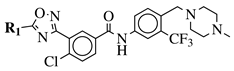 |
R1 = 3-pyridinyl, 4-pyridinyl, 4-methoxy-3-pyridinyl, 5-(2-methoxyethoxy)-3-pyridinyl, 5-morpholin-3-pyridinyl or 5-(1-methyl-1H-pyrazol-3-yl)-3-pyridinyl. | 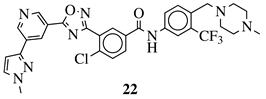 |
IC50 value of 7.3 nM against RET enzyme in ELISA assay. | [57] |
4. Antimicrobial Agents
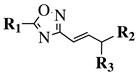 | ||||
| R | ||||
| 1 | ||||
| = phenyl, 4-mehtoxyphenyl, 4-chlorophenyl, 3-methylthienyl or 2-pyridinyl; | ||||
| R2, R3 = H, methyl, phenyl, 4-chlorophenyl, 4-methoxyphenyl, 3,4,-dimethoxyphenyl or 2,3-dimethoxyphenyl. |  |
MIC value of 0.68 mM against S. aureus. | [108][77] | |
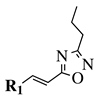 |
R1 = phenyl, 4-methylphenyl, 4-methoxyphenyl, 4-methylthiophenyl, 2-chlorophenyl, 4-chlorophenyl, 2-3-dichlorophenyl, 3,4-dichlorophenyl, 4-fluorophenyl, 4-bromophenyl, 4-hydroksyphenyl, 2-bromo-4-fluorophenyl, 4-cyanophenyl, 4-pyridinyl, 1-napthyl and others (see Ref.). | 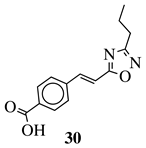 |
IC50 value of 0.045 μg/mL against M. tuberculosis (H37Ra). | [109][78] |
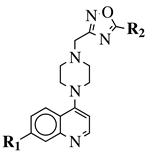 |
R1 = 4-pyridyl, 3-pyridinyl or 3,5-difluorophenyl; R2 = 3,5-dimethoxyphenyl, 3,5-difluorophenyl, 3-cyanophenyl, 2,3-dimethylphenyl, cyclopentyl or 4-izopropylphenyl. |
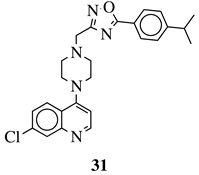 |
MIC value of 0.5 μg/mL against M. tuberculosis (H37Ra). | [110][79] |
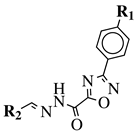 |
R1 = H, F, Cl, Br, methyl, nitro, methoxy or hydroxy; R2 = 4-hydroksy-3-methoxyphenyl, 2-styryl, ferrocene or 5-benzo[1,3]dioxole. |
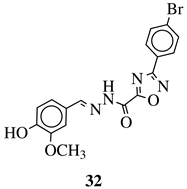 |
IC50 value of 0.02 μM against P. falciparum. In vivo studies failed—none in vivo activity. |
[111][80] |
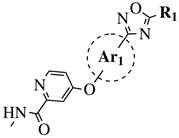 |
R1 = Me, Et, cyclopropyl, iPr, CF3, iBu or CH2OCH3; Ar1 = p-phenylene, p-2-methylphenylene, p-2,6-dimethylphenylene, 2,5-pyridinyl or 3-methylbenzothiophene |
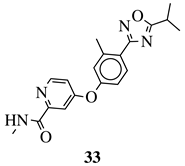 |
IC50 values of 66.0, 22.0 and 3.7 nM against hRV-B14, hRV-A21 and hRV-A71, respectively. | [112][81] |
References
- Salahuddin; Mazumder, A.; Yar, M.S.; Mazumder, R.; Chakraborthy, G.S.; Ahsan, M.J.; Rahman, M.U. Updates on synthesis and biological activities of 1,3,4-oxadiazole: A review. Synth. Commun. 2017, 47, 1805–1847.
- Bajaj, S.; Asati, V.; Singh, J.; Roy, P.P. 1,3,4-Oxadiazoles: An emerging scaffold to target growth factors, enzymes and kinases as anticancer agents. Eur. J. Med. Chem. 2015, 97, 124–141.
- Bala, S.; Saini, V.; Kamboj, S.; Prasad, D.N. Review Exploring Antiinflammatory Potential of 1,3,4-Oxadiazole Derivatives as Promising Lead. Int. J. Pharm. Sci. Rev. Res. 2012, 17, 84–89.
- Khalilullah, H.J.; Ahsan, M.; Hedaitullah, M.; Khan, S.; Ahmed, B. 1,3,4-Oxadiazole: A Biologically Active Scaffold. Mini-Rev. Med. Chem. 2012, 12, 789–801.
- Bajaj, S.; Roy, P.P.; Singh, J. 1,3,4-Oxadiazoles as Telomerase Inhibitor: Potential Anticancer Agents. Anti-Cancer Agents Med. Chem. 2018, 17, 1869–1883.
- WebOfScience. Available online: (accessed on 16 December 2019).
- Wei, H.; He, C.; Zhang, J.; Shreeve, J.M. Combination of 1,2,4-Oxadiazole and 1,2,5-Oxadiazole Moieties for the Generation of High-Performance Energetic Materials. Angew. Chem. 2015, 127, 9499–9503.
- Boiani, M. 1,2,5-Oxadiazole N-oxide derivatives as potential anti-cancer agents: Synthesis and biological evaluation. Part IV. Eur. J. Med. Chem. 2001, 36, 771–782.
- Fershtat, L.L.; Makhova, N.N. 1,2,5-Oxadiazole-Based High-Energy-Density Materials: Synthesis and Performance. ChemPlusChem 2020, 85, 13–42.
- Nguyen, M.T.; Hegarty, A.F.; Elguero, J. Can 1,2,3-Oxadiazole be Stable? Angew. Chem. Int. Ed. Engl. 1985, 24, 713–715.
- Tiemann, F.; Krüger, P. Ueber Amidoxime und Azoxime. Berichte Der Dtsch. Chem. Ges. 1884, 17, 1685–1698.
- Newman, H. Photochemistry of 3,5-diphenyl-1,2,4-oxadiazole II. Photolysis in protic media. Tetrahedron Lett. 1968, 9, 2421–2424.
- Newman, H. Photochemistry of 3,5-diphenyl-1,2,4-oxadiazole I. Photolysis in aprotic media. Tetrahedron Lett. 1968, 9, 2417–2420.
- Anderson, G.W.; Faith, H.E.; Marson, H.W.; Winnek, P.S.; Roblin, R.O. Studies in Chemotherapy. VI. Sulfanilamido Heterocycles. J. Am. Chem. Soc. 1942, 64, 2902–2905.
- Silvestrini, B.; Catanese, B. Ricerche sul metabolismo del 5-beta-dietilamino-3-alfa -fenilpropil-1,2,4-oxadiazolo. Bollettino Chimico Farmaceutico 1964, 103, 447–450.
- Silvestrini, B. Un antitosse-antinfiammatorio, l’Oxolamina (Perebron). Minerva Medica 1960, 51, 4091–4094.
- Parra, M.; Hidalgo, P.; Alderete, J. New supramolecular liquid crystals induced by hydrogen bonding between pyridyl-1,2,4-oxadiazole derivatives and 2,5-thiophene dicarboxylic acid. Liq. Cryst. 2005, 32, 449–455.
- Xiong, H.; Yang, H.; Lei, C.; Yang, P.; Hu, W.; Cheng, G. Combinations of furoxan and 1,2,4-oxadiazole for the generation of high performance energetic materials. Dalton Trans. 2019, 48, 14705–14711.
- Yan, T.; Cheng, G.; Yang, H. 1,2,4-Oxadiazole-Bridged Polynitropyrazole Energetic Materials with Enhanced Thermal Stability and Low Sensitivity. ChemPlusChem 2019, 84, 1567–1577.
- Pitasse-Santos, P.; Sueth-Santiago, V.; Lima, M. 1,2,4- and 1,3,4-Oxadiazoles as Scaffolds in the Development of Antiparasitic Agents. J. Braz. Chem. Soc. 2017, 29, 435–456.
- Rosa, M.F.; Morcelli, A.C.T.; Lobo, V.S. 1,2,4-Oxadiazole: A Brief Review From The Literature About the Synthesis and Pharmacological Applications. Vis ao Acadêmica Curitiba 2015, 16, 130–157.
- Coupar, I.M.; Hedges, A.; Metcalfe, H.L.; Turner, P. Effect of aminophylline, butalamine and imolamine on human isolated smooth muscle. J. Pharm. Pharmacol. 1969, 21, 474–475.
- Rotbart, H.A.; Webster, A.D. Treatment of Potentially Life-Threatening Enterovirus Infections with Pleconaril. Clin. Infect. Dis. 2001, 32, 228–235.
- McDonald, C.M.; Campbell, C.; Torricelli, R.E.; Finkel, R.S.; Flanigan, K.M.; Goemans, N.; Heydemann, P.; Kaminska, A.; Kirschner, J.; Muntoni, F.; et al. Ataluren in patients with nonsense mutation Duchenne muscular dystrophy (ACT DMD): A multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1489–1498.
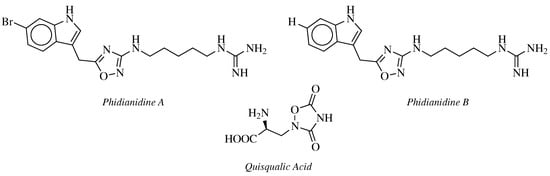
- Carbone, M.; Li, Y.; Irace, C.; Mollo, E.; Castelluccio, F.; Di Pascale, A.; Cimino, G.; Santamaria, R.; Guo, Y.W.; Gavagnin, M. Structure and Cytotoxicity of Phidianidines A and B: First Finding of 1,2,4-Oxadiazole System in a Marine Natural Product. Org. Lett. 2011, 13, 2516–2519.
- Vitale, R.M.; Gatti, M.; Carbone, M.; Barbieri, F.; Felicità, V.; Gavagnin, M.; Florio, T.; Amodeo, P. A minimalist hybrid ligand/receptor-based pharmacophore model for CXCR4 applied to a small-library of marine natural products led to the identification of Phidianidine A as a new CXCR4 ligand exhibiting antagonist activity. ACS Chem. Biol. 2013, 8, 2762–2770.
- Zhang, L.; Jiang, C.S.; Gao, L.X.; Gong, J.X.; Wang, Z.H.; Li, J.Y.; Li, J.; Li, X.W.; Guo, Y.W. Design, synthesis and in vitro activity of phidianidine B derivatives as novel PTP1B inhibitors with specific selectivity. Bioorg. Med. Chem. Lett. 2016, 26, 778–781.
- Hermit, M.B.; Greenwood, J.R.; Bräuner-Osborne, H. Mutation-induced Quisqualic Acid and Ibotenic Acid Affinity at the Metabotropic Glutamate Receptor Subtype 4. J. Biol. Chem. 2004, 279, 34811–34817.
- Kozikowski, A.P.; Steensma, D.; Varasi, M.; Pshenichkin, S.; Surina, E.; Wroblewski, J.T. α-substituted quisqualic acid analogs: New metabotropic glutamate receptor group II selective antagonists. Bioorg. Med. Chem. Lett. 1998, 8, 447–452.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Zhang, H.Z.; Kasibhatla, S.; Kuemmerle, J.; Kemnitzer, W.; Ollis-Mason, K.; Qiu, L.; Crogan-Grundy, C.; Tseng, B.; Drewe, J.; Cai, S.X. Discovery and Structure-Activity Relationship of 3-Aryl-5-aryl-1,2,4-oxadiazoles as a New Series of Apoptosis Inducers and Potential Anticancer Agents. J. Med. Chem. 2005, 48, 5215–5223.
- Pace, A.; Buscemi, S.; Piccionello, A.P.; Pibiri, I. Recent Advances in the Chemistry of 1,2,4-Oxadiazoles. In Advances in Heterocyclic Chemistry; Academic Press Inc.: Cambridge, MA, USA, 2015; Volume 116, pp. 85–136.
- Rasool, I.; Ahmad, M.; Khan, Z.A.; Mansha, A.; Maqbool, T.; Zahoor, A.F.; Aslam, S. Recent advancements in oxadiazole-based anticancer agents. Trop. J. Pharm. Res. 2017, 16, 723.
- Maftei, C.V.; Fodor, E.; Jones, P.G.; Franz, M.H.; Kelter, G.; Fiebig, H.; Neda, I. Synthesis and characterization of novel bioactive 1,2,4-oxadiazole natural product analogs bearing the N-phenylmaleimide and N-phenylsuccinimide moieties. Beilstein J. Org. Chem. 2013, 9, 2202–2215.
- Maftei, C.V.; Fodor, E.; Jones, P.G.; Daniliuc, C.G.; Franz, M.H.; Kelter, G.; Fiebig, H.H.; Tamm, M.; Neda, I. Novel 1,2,4-oxadiazoles and trifluoromethylpyridines related to natural products: Synthesis, structural analysis and investigation of their antitumor activity. Tetrahedron 2016, 72, 1185–1199.
- Maftei, C.V.; Fodor, E.; Jones, P.G.; Freytag, M.; Franz, M.H.; Kelter, G.; Fiebig, H.H.; Tamm, M.; Neda, I. N -heterocyclic carbenes (NHC) with 1,2,4-oxadiazole-substituents related to natural products: Synthesis, structure and potential antitumor activity of some corresponding gold(I) and silver(I) complexes. Eur. J. Med. Chem. 2015, 101, 431–441.
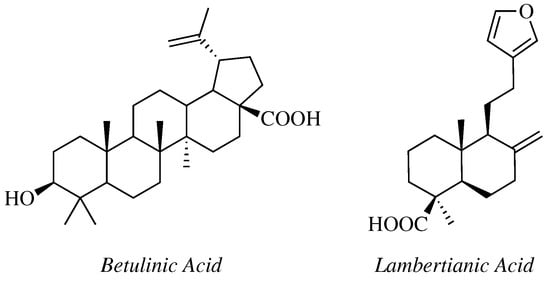
- Krishna, C.; Bhargavi, M.V.; Krupadanam, G.L.D. Design, Synthesis, and Cytotoxicity of Semisynthetic Betulinic Acid-1,2,4-Oxadiazole Amide Derivatives. Russ. J. Gen. Chem. 2018, 88, 312–318.
- Challa, K.; Bhargavi, M.V.; Krupadanam, G.L.D. Design, semisynthesis and cytotoxic activity of novel ester derivatives of betulinic acid-1,2,4 oxadiazoles. J. Asian Nat. Prod. Res. 2016, 18, 1158–1168.
- Mironov, M.E.; Pokrovsky, M.A.; Kharitonov, Y.V.; Shakirov, M.M.; Pokrovsky, A.G.; Shults, E.E. Furanolabdanoid-based 1,2,4-oxadiazoles: Synthesis and cytotoxic activity. ChemistrySelect 2016, 1, 417–424.
- Kucukoglu, K.; Tugrak, M.; Demirtas, A.; Sakagami, H.; Gul, H.I. Synthesis and Cytotoxic Activity of (4-Substituted-benzylidene)-(3-Phenyl-1,2,4-Oxadiazol-5-YL)Methylamines. Pharm. Chem. J. 2016, 50, 234–238.
- Moniot, S.; Forgione, M.; Lucidi, A.; Hailu, G.S.; Nebbioso, A.; Carafa, V.; Baratta, F.; Altucci, L.; Giacché, N.; Passeri, D.; et al. Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure–Activity Relationship, X-ray Crystal Structure, and Anticancer Activity. J. Med. Chem. 2017, 60, 2344–2360.
- Avanzo, R.E.; Padrón, J.M.; D’Accorso, N.B.; Fascio, M.L. Synthesis and in vitro antiproliferative activities of (5-aryl-1,2,4-oxadiazole-3-yl) methyl D-ribofuranosides. Bioorg. Med. Chem. Lett. 2017, 27, 3674–3677.
- Abd el hameid, M.K.; Mohammed, M.R. Design, synthesis, and cytotoxicity screening of 5-aryl-3-(2-(pyrrolyl) thiophenyl)-1, 2, 4-oxadiazoles as potential antitumor molecules on breast cancer MCF-7 cells. Bioorg. Chem. 2019, 86, 609–623.
- de Oliveira, V.N.M.; dos Santos, F.G.; Ferreira, V.P.G.; Araújo, H.M.; do Ó Pessoa, C.; Nicolete, R.; de Oliveira, R.N. Focused microwave irradiation-assisted synthesis of N-cyclohexyl-1,2,4-oxadiazole derivatives with antitumor activity. Synth. Commun. 2018, 48, 2522–2532.
- Sateesh Kumar, P.; Umadevi, P. Novel Bis(1,2,4-oxadiazolyl) Fused Thiazole Derivatives: Synthesis and Anticancer Activity. Russ. J. Gen. Chem. 2018, 88, 2611–2615.
- Pervaram, S.; Ashok, D.; Sarasija, M.; Reddy, C.V.R.; Sridhar, G. Synthesis and Anticancer Activity of 1,2,4-Oxadiazole Fused Benzofuran Derivatives. Russ. J. Gen. Chem. 2018, 88, 1219–1223.
- Chakrapani, B.; Ramesh, V.; Pourna Chander Rao, G.; Ramachandran, D.; Madhukar Reddy, T.; Kalyan Chakravarthy, A.; Sridhar, G. Synthesis and Anticancer Evaluation of 1,2,4-Oxadiazole Linked Imidazothiadiazole Derivatives. Russ. J. Gen. Chem. 2018, 88, 1020–1024.
- Srinivas, M.; Satyaveni, S.; Ram, B. Synthesis and Anticancer Activity of 1,2,4-Oxadiazol Linked Benzimidazole Derivatives. Russ. J. Gen. Chem. 2018, 88, 2653–2657.
- Chiacchio, M.A.; Legnani, L.; Campisi, A.; Paola, B.; Giuseppe, L.; Iannazzo, D.; Veltri, L.; Giofrè, S.; Romeo, R. 1,2,4-Oxadiazole-5-ones as analogues of tamoxifen: Synthesis and biological evaluation. Org. Biomol. Chem. 2019, 17, 4892–4905.
- Krasavin, M.; Shetnev, A.; Sharonova, T.; Baykov, S.; Tuccinardi, T.; Kalinin, S.; Angeli, A.; Supuran, C.T. Heterocyclic periphery in the design of carbonic anhydrase inhibitors: 1,2,4-Oxadiazol-5-yl benzenesulfonamides as potent and selective inhibitors of cytosolic hCA II and membrane-bound hCA IX isoforms. Bioorg. Chem. 2018, 76, 88–97.
- Krasavin, M.; Shetnev, A.; Sharonova, T.; Baykov, S.; Kalinin, S.; Nocentini, A.; Sharoyko, V.; Poli, G.; Tuccinardi, T.; Presnukhina, S.; et al. Continued exploration of 1,2,4-oxadiazole periphery for carbonic anhydrase-targeting primary arene sulfonamides: Discovery of subnanomolar inhibitors of membrane-bound hCA IX isoform that selectively kill cancer cells in hypoxic environment. Eur. J. Med. Chem. 2019, 164, 92–105.
- Cascioferro, S.; Attanzio, A.; Di Sarno, V.; Musella, S.; Tesoriere, L.; Cirrincione, G.; Diana, P.; Parrino, B. New 1,2,4-Oxadiazole Nortopsentin Derivatives with Cytotoxic Activity. Mar. Drugs 2019, 17, 35.
- Polothi, R.; Raolji, G.S.B.; Kuchibhotla, V.S.; Sheelam, K.; Tuniki, B.; Thodupunuri, P. Synthesis and biological evaluation of 1,2,4-oxadiazole linked 1,3,4-oxadiazole derivatives as tubulin binding agents. Synth. Commun. 2019, 49, 1603–1612.
- Yang, F.; Shan, P.; Zhao, N.; Ge, D.; Zhu, K.; Jiang, C.s.; Li, P.; Zhang, H. Development of hydroxamate-based histone deacetylase inhibitors containing 1,2,4-oxadiazole moiety core with antitumor activities. Bioorg. Med. Chem. Lett. 2019, 29, 15–21.
- Yang, F.; Zhang, T.; Wu, H.; Yang, Y.; Liu, N.; Chen, A.; Li, Q.; Li, J.; Qin, L.; Jiang, B.; et al. Design and Optimization of Novel Hydroxamate-Based Histone Deacetylase Inhibitors of Bis-Substituted Aromatic Amides Bearing Potent Activities against Tumor Growth and Metastasis. J. Med. Chem. 2014, 57, 9357–9369.
- Yang, Z.; Shen, M.; Tang, M.; Zhang, W.; Cui, X.; Zhang, Z.; Pei, H.; Li, Y.; Hu, M.; Bai, P.; et al. Discovery of 1,2,4-oxadiazole-Containing hydroxamic acid derivatives as histone deacetylase inhibitors potential application in cancer therapy. Eur. J. Med. Chem. 2019, 178, 116–130.
- Han, M.; Li, S.; Ai, J.; Sheng, R.; Hu, Y.; Hu, Y.; Geng, M. Discovery of 4-chloro-3-(5-(pyridin-3-yl)-1,2,4-oxadiazole-3-yl)benzamides as novel RET kinase inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 5679–5684.
- Hande, K. Etoposide: Four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 1998, 34, 1514–1521.
- Guest, J.F.; Panca, M.; Sladkevicius, E.; Gough, N.; Linch, M. Cost Effectiveness of First-Line Treatment with Doxorubicin/Ifosfamide Compared to Trabectedin Monotherapy in the Management of Advanced Soft Tissue Sarcoma in Italy, Spain, and Sweden. Sarcoma 2013, 2013, 1–19.
- Carvalho, C.; Santos, R.; Cardoso, S.; Correia, S.; Oliveira, P.; Santos, M.; Moreira, P. Doxorubicin: The Good, the Bad and the Ugly Effect. Curr. Med. Chem. 2009, 16, 3267–3285.
- Avanzo, R.E.; Anesini, C.; Fascio, M.L.; Errea, M.I.; D’Accorso, N.B. 1,2,4-Triazole D-ribose derivatives: Design, synthesis and antitumoral evaluation. Eur. J. Med. Chem. 2012, 47, 104–110.
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host Range and Emerging and Reemerging Pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847.
- Dye, C. After 2015: Infectious diseases in a new era of health and development. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130426.
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128.
- O’Daniel, P.I.; Peng, Z.; Pi, H.; Testero, S.A.; Ding, D.; Spink, E.; Leemans, E.; Boudreau, M.A.; Yamaguchi, T.; Schroeder, V.A.; et al. Discovery of a New Class of Non-β-lactam Inhibitors of Penicillin-Binding Proteins with Gram-Positive Antibacterial Activity. J. Am. Chem. Soc. 2014, 136, 3664–3672.
- Carter, G.P.; Harjani, J.R.; Li, L.; Pitcher, N.P.; Nong, Y.; Riley, T.V.; Williamson, D.A.; Stinear, T.P.; Baell, J.B.; Howden, B.P. 1,2,4-Oxadiazole antimicrobials act synergistically with daptomycin and display rapid kill kinetics against MDR Enterococcus faecium. J. Antimicrob. Chemother. 2018, 73, 1562–1569.
- Ding, D.; Boudreau, M.A.; Leemans, E.; Spink, E.; Yamaguchi, T.; Testero, S.A.; O’Daniel, P.I.; Lastochkin, E.; Chang, M.; Mobashery, S. Exploration of the structure–activity relationship of 1,2,4-oxadiazole antibiotics. Bioorg. Med. Chem. Lett. 2015, 25, 4854–4857.
- Spink, E.; Ding, D.; Peng, Z.; Boudreau, M.A.; Leemans, E.; Lastochkin, E.; Song, W.; Lichtenwalter, K.; O’Daniel, P.I.; Testero, S.A.; et al. Structure–Activity Relationship for the Oxadiazole Class of Antibiotics. J. Med. Chem. 2015, 58, 1380–1389.
- Leemans, E.; Mahasenan, K.V.; Kumarasiri, M.; Spink, E.; Ding, D.; O’Daniel, P.I.; Boudreau, M.A.; Lastochkin, E.; Testero, S.A.; Yamaguchi, T.; et al. Three-Dimensional QSAR Analysis and Design of New 1,2,4-Oxadiazole Antibacterials. Bioorg. Med. Chem. Lett. 2016, 26, 1011–1015.
- Xiao, Q.; Vakulenko, S.; Chang, M.; Mobashery, S. Mutations in mmpL and in the Cell Wall Stress Stimulon Contribute to Resistance to Oxadiazole Antibiotics in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 5841–5847.
- Janardhanan, J.; Chang, M.; Mobashery, S. The oxadiazole antibacterials. Curr. Opin. Microbiol. 2016, 33, 13–17.
- Ceballos, S.; Kim, C.; Ding, D.; Mobashery, S.; Chang, M.; Torres, C. Activities of Oxadiazole Antibacterials against Staphylococcus aureus and Other Gram-Positive Bacteria. Antimicrob. Agents Chemother. 2018, 62.
- Krolenko, K.Y.; Vlasov, S.V.; Zhuravel, I.A. Synthesis and antimicrobial activity of 5-(1H-1,2,3-triazol-4-yl)-1,2,4-oxadiazole derivatives. Chem. Heterocycl. Compd. 2016, 52, 823–830.
- Cunha, F.; Nogueira, J.; de Aguiar, A. Synthesis and Antibacterial Evaluation of 3,5-Diaryl-1,2,4-oxadiazole Derivatives. J. Braz. Chem. Soc. 2018, 29, 2405–2416.
- Shi, G.; He, X.; Shang, Y.; Xiang, L.; Yang, C.; Han, G.; Du, B. Synthesis of 3′,4′-Diaryl-4′H -spiro[indoline-3,5′-[1′,2′,4′]oxadiazol]-2-ones via DMAP-catalyzed Domino Reactions and Their Antibacterial Activity. Chin. J. Chem. 2016, 34, 901–909.
- Shetnev, A.; Baykov, S.; Kalinin, S.; Belova, A.; Sharoyko, V.; Rozhkov, A.; Zelenkov, L.; Tarasenko, M.; Sadykov, E.; Korsakov, M.; et al. 1,2,4-Oxadiazole/2-Imidazoline Hybrids: Multi-target-directed Compounds for the Treatment of Infectious Diseases and Cancer. Int. J. Mol. Sci. 2019, 20, 1699.
- Tarasenko, M.; Sidneva, V.; Belova, A.; Romanycheva, A.; Sharonova, T.; Baykov, S.; Shetnev, A.; Kofanov, E.; Kuznetsov, M. An efficient synthesis and antimicrobial evaluation of 5-alkenyl- and 5-styryl-1,2,4-oxadiazoles. Arkivoc 2018, 2018, 458–470.
- Atmaram Upare, A.; Gadekar, P.K.; Sivaramakrishnan, H.; Naik, N.; Khedkar, V.M.; Sarkar, D.; Choudhari, A.; Mohana Roopan, S. Design, synthesis and biological evaluation of (E)-5-styryl-1,2,4-oxadiazoles as anti-tubercular agents. Bioorg. Chem. 2019, 86, 507–512.
- Shruthi, T.; Eswaran, S.; Shivarudraiah, P.; Narayanan, S.; Subramanian, S. Synthesis, antituberculosis studies and biological evaluation of new quinoline derivatives carrying 1,2,4-oxadiazole moiety. Bioorg. Med. Chem. Lett. 2019, 29, 97–102.
- dos Santos Filho, J.M.; de Queiroz e Silva, D.M.A.; Macedo, T.S.; Teixeira, H.M.P.; Moreira, D.R.M.; Challal, S.; Wolfender, J.L.; Queiroz, E.F.; Soares, M.B.P. Conjugation of N-acylhydrazone and 1,2,4-oxadiazole leads to the identification of active antimalarial agents. Bioorg. Med. Chem. 2016, 24, 5693–5701.
- Kim, J.; Shin, J.S.; Ahn, S.; Han, S.B.; Jung, Y.S. 3-Aryl-1,2,4-oxadiazole Derivatives Active Against Human Rhinovirus. ACS Med. Chem. Lett. 2018, 9, 667–672.
