COVID-19 viral disease is officially global pandemic, currently accounting for the highest number of deaths worldwide. Special screening is extremely important as an effective way to monitor and manage the pandemic before reaching herb immunity through effective vaccination against SARS-CoV-2. A rapid population control task for COVID-19 has been documented using innovative methods in biosensor development. Biosensors are selected as promising detection devices with enormous potential as point-of-care (POC) tools to confirm the SARS-CoV-2 infection. Timely testing also helps to effectively allocate medical resources and save time for frontline medical staff. Hence, simple, rapid, cost-effective, and accessible detection techniques as POC diagnostics for large-scale screening and field testing of SARS-CoV-2 infection is important and should urgently be expedited to control the rapid and contagious spread of COVID-19.
- SARS-CoV-2
- COVID-19 clinical diagnostics
- nucleic acid amplification
- RT-PCR
- RT-LAMP
- optical biosensor
- lateral flow assay
- ELISA
- electrochemical biosensor
- lab-in-a-tube
1. Nucleic Acid Amplification-Based Techniques as Gold Standard Diagnostic Tests
A variety of NAA techniques have been incorporated into well-known clinical diagnostic tests, such as polymerase chain reaction (PCR), real-time PCR, and reverse transcription-mediated isothermal amplification (RT-LAMP). These techniques have widespread research applications in the diagnosis of COVID-19 as the gold standard diagnosis at the start of a pandemic. These techniques are well-developed owing to their simplicity, sensitivity, and speed. Chu et al. 2020 developed a one-step quantitative real-time RT-PCR in addition to a biological sensor for monitoring two different regions (ORF1b and ORFN) of the SARS-CoV-2 viral genome. Instead of two-step RT-PCR that separately conducts RNA reverse transcription and amplification steps, reverse transcriptase and DNA polymerase enzymes were premixed in a single tube that allows both steps to be performed in single reaction. These NAA-based sensors excel in the analytical requirements but are typically limited due to their time-consuming processes. RT-PCR platform has a lengthy laboratory workflow requiring multiple solution operation steps and relies on sophisticated equipment for thermal cycling and optical signal detection. It may not be a viable option for the screening of COVID-19 at locations where laboratories and highly trained technicians are absent. The aim of one-step quantitative RT-PCR is the rapid detection of SARS-CoV-2 in human samples (75 min), which is highly acceptable in clinical tests, however, it is more cost-effective, and required a robust manner in laboratories in different geographical regions. This two-stage amplification closed-tube diagnostic assay was used to test SARS-CoV-2 samples with significantly enhanced sensitivity compared to conventional RT-PCR with dynamic range of at least seven orders of magnitude (2 × 10−4–2000 TCID50/reaction) for RNA from SARS infected cells and below 10 copies per reaction for DNA plasmid as positive standards [1]. Chow et al. 2020 demonstrated the potential of the LAMP-based sensor of SARS-CoV-2 by monitoring the color change of the different concentrations of clinical samples which was detectable by naked eye [2]. This method was designed to directly amplify the target through one-step RT-LAMP test, then target RNA was confirmed via colorimetric method that showed the efficiency of detection of target RNA genome within range of 45–105 min including sample extraction time, depending on viral load. The tested clinical samples, which were different in nature (respiratory samples, nasopharyngeal swabs, sputum/deep throat saliva, and throat swabs), were confirmed by RT-PCR and collected at different reaction times (60 min and 90 min). Interestingly, the test with nasopharyngeal swab samples showed the highest sensitivity, with 96.88% (95% CI: 0.93–1.00) and 98.96% (95% CI: 0.97–1.00) of 96 samples positive by RT-LAMP at 60 and 90 min, respectively. In addition, the other clinical samples had high sensitivity of at least 93.33% (0.87–1.00), suggesting that this platform exhibits high potential in the detection application of SARS-CoV-2. In addition, Viet Loan et al. 2020 investigated key issues related to the colorimetric RT-LAMP assay and LAMP-sequencing, sensor characteristics, sensitivity and specificity in detecting SARS-CoV-2 RNA both in vitro and in vivo. The inclusion of RT-LAMP and colorimetric methods in the matrix increased the simple, scalable, and broadly applicable testing methods of the sensor. Furthermore, the sensor characteristics of the colorimetric RT-LAMP assay were tested on 768 pharyngeal swab specimens using a primer set specific for the N gene and compared an RT-PCR assay using a sensitive primer set with a sensitivity of 97.5% and specificity of 99.7%. In particular, the swab-to-RT-LAMP assay without a prior RNA isolation step showed excellent specificity (99.5%) but lower sensitivity (86% for CT < 30) than the RT-LAMP assay [3] (Figure 1). Furthermore, Ackerman et al. 2020 developed combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) for the evaluation of multiplex pathogenic nucleic acids to detect pathogens. CRISPR-based nucleic acid detection reagents containing in nanoliter droplets could self-organize in arrays to pair with droplets of amplifies samples, testing samples against CRISPR RNA (crRNA) in replicate. With the combination of CARMEN and Cas13 detection, the assay simultaneously differentiated 169 human-related viruses and incorporated an additional crRNA to detect target of COVID-19. The CARMEN assay enables the scalability, miniaturization, and cost-effectiveness, shifting diagnostic ability from targeted high-priority samples to comprehensive large samples [4]. Traditional NAA technique is time-consuming and may have false-positive outputs based on the working experience of the technician with careful consideration throughout the testing time. However, PCR has become an indispensable and integral part of clinical and diagnostic research as the gold standard in hospitals due to their unique performance. One of the significant applications of the NAA-based techniques was their utility in the confirmation of SARS-CoV-2 infection for the better intervention of COVID-19 pandemic, which can shift diagnostic and surveillance efforts from targeted testing of high-priority samples to comprehensive testing of large sample sets, bringing great benefits to patients and public health.
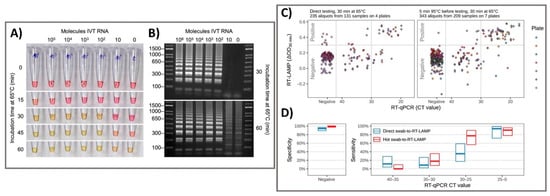
Figure 1. Colorimetric RT-LAMP and LAMP sequencing for the clinical detection of SARS-CoV-2 RNA. (A) The oligonucleotide set for the nucleocapsid (N) gene of SARS-CoV-2 was added to the RT-LAMP reaction and incubated at 65 °C. The colors of samples changed from red-to-yellow and the negative control was yellowish. (B) Gel electrophoresis showed RT-LAMP reaction products with distinct banding patterns. (C) For clinical pharyngeal swab samples, the direct swab-to-RT-LAMP assay measurements or after 5 min of heat treatment at 95 °C were compared for their ΔOD values from the swab-to-RT-LAMP assay and CT values from the RT-qPCR assay. (D) The sensitivity and specificity of the swab-to-RT-LAMP assay were revealed with their 95% confidence intervals, with the direct swab-to-RT-LAMP assay (blue color) and the heated swab-to-RT-LAMP assay (red color). Reprinted with permission from [3]. Sci. Transl. Med. 2020, 12, 556, eabc7075. Copyright 2020, American Association for the Advancement of Science.
2. Optical Sensing Platforms as Rapid Point-Of-Care Screening Tests
Optical biosensors are one of the most common platforms that have been exploited to monitor various target for clinical diagnostics. They detect biological interactions by evaluating induced variations in the properties of light, such as intensity, wavelength, index of refraction, or polarization. Cutting-edge optical sensing platform technologies are currently being investigated for COVID-19 clinical samples, including samples based on lateral flow assays (LFAs), enzyme-linked immunosorbent assay (ELISA), chemiluminescent immunoassay, plasmonic biosensor, and localized surface plasmon resonance. Grant et al. 2020 developed half-strip LFAs as useful first step in the development of LFA platforms for the detection of SARS-CoV-2 using commercially available antibodies. This half-strip LFA exhibited high sensitivity toward SARS-CoV-2 with limit of detection (LOD) at 0.65 ng/mL by visual read or optical reader [5] (Figure 2). LFA is a good biosensing candidate for diagnostic applications owing to their advantages including high sensitivity and specificity, excellent biological compatibility, short duration, stable output, and affordability. More importantly, the LFA holds the potential for large-scale production and commercialization with convenient protocol without technical professions, allowing it to be POC test worldwide for initial screening of SARS-CoV-2 infection. Due to the feasibility of LFA biosensing platforms for the effective detection of SARS-CoV-2 biomarkers, there were several LFA strips developed from different companies. Demey et al. 2020 evaluated the sensing performance to detect SARS-CoV-2 using four immunochromatographic antibody assays of different commercial companies. Using SARS-CoV-2 positive samples confirmed by RT-PCR from 22 patients, they demonstrated that the ability of COVID-19 confirmation through antibody using these LFAs was depended on time with the median detection time about 8–10 days since the onset of symptoms, and the sensitivity was increased up to 60–80% on day 10 and 100% on day 15, indicating that these LFA tests were reliable at 14–15 days post-infection [6]. The low-cost LFA combined with an easily accessible synthetic biosensor that can function with bodily fluids as samples provides a comprehensive solution for the diagnosis of non-communicable diseases in resource-limited developing countries [7]. Instead of LFAs, the use of dual-functional plasmonic biosensor by combining plasmonic photothermal and localized surface plasmon resonance sensing transduction could provide a promising alternative optical biosensor. Qiu et al. 2020 identified an optical LOD of approximately 0.22 pM and confirmed a detection range of 0.01 pM to 50 μM for the detection of target SARS-CoV-2 sequences [8]. The highly sensitivive detection of target sequences was achieved by using 2D gold nanoislands (AuNIs) functionalized with complementary DNA receptors through nucleic acid hybridization. To enhance sensing performance, thermoplasmonic heat was generated on AuNI chip under illuminated at plasmonic resonance frequencies, elevating the in situ hybridization temperature and facilitating the accurate differentiation of two similar gene sequences. This dual-functional plasmonic biosensor exhibited the potential application in nucleic acid tests for viral disease diagnosis. Overall, these types of optical sensing platform enable the detection of SARS-CoV-2 biomarkers for the confirmation of COVID-19 in human samples including serum, plasma, blood, nasopharyngeal, and oropharyngeal swab specimens, with high sensitivity, specificity. With these advantages, the strong capability of translation of the signal intensity into the accurate concentration of biomarkers makes optical platforms important as applicable biosensors for POC diagnostic of COVID-19 in clinical. Therefore, almost clinical tests utilized optical sensing platform have been currently exploited as an effective tool for diagnosis of disease by sensing of biomarkers in human biological specimens. Compared to other optical biosensors that have been developed to detect SARS-CoV-2, it is worth noticing that LFA exhibits the superior potential to serve as diagnostic tools for initial screening of COVID-19 with acceptable results, short-time consumption and reasonable price, suggesting a good choice for developing and underdeveloped countries to utilize LFAs as POC diagnostic tool for managing the COVID-19 pandemic.
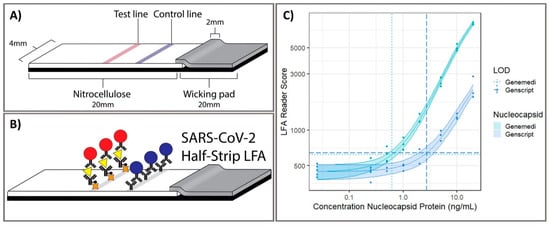
Figure 32. Point-of-care half-strip lateral flow assay for the detection of the nucleocapsid antigen of SARS-CoV-2. (A) The 4 mm width half-strip was constructed using a 20 mm nitrocellulose analytical membrane, 20 mm wicking pad by using a Kinematic Matrix guillotine cutter. (B) LFA was treated in buffer and color intensity of test zone was differentiated after 20 min. (C) The dosage response curve for half-band LFA using nucleocapsid proteins from two commercially available sources, measured with commercially available optical LFA readers. Reprinted with permission from [5]. Anal. Chem. 2020, 92, 16, 11305–11309. Copyright 2020, American Chemical Society.
3. Lab-In-A-Tube and Electrochemical Sensors as Emerging Ultrasensitive Real-Time Monitors
Laboratory diagnostics which compatible with critical laboratory equipment are conceptually easily applied to patients, given the features of the following multiple operations of virus testing [9]. Exposure tracking can limit the viral spread, however, population screening to determine virus infection levels in the community is a longer-term need. For lab-in-a-tube, Alves et al. 2020 built a sensing platform using gel tag agglutination tests to target SARS-CoV-2 with rapid case identification (Figure 3). Ten serological samples in both gel cards and indirect IgG ELISA were tested and showed that similar performance between them, suggesting this assay as one of suitable approach for clinical diagnosis compared to conventional ELISA, owing to its advantages in excellent resolution and benefits of high throughput, high speed (10–30 min), automatic in most cases, and possibility for POC diagnostics. From gel card agglutination assays to lab-in-a-tube systems, this simple, rapid, and scalable approach could identify disease-specific activity to apply in SARS-CoV-2 testing, suggesting the ability to move diagnostics to the POC test for COVID-19 confirmation [10]. In addition, Lin et al. 2020 recently demonstrated real-time and continuous monitoring platform using integrated diagnostic microchips, homemade mobile fluorescence detectors, and microfluidic immunoassay systems for the simultaneous detection of IgG/IgM/antigen in SARS-CoV-2. This system was utilized for SARS-CoV-2 serological testing, displaying high accuracy not only in distinguishing between infected and uninfected cases but also in determining the severity the disease, allowing disease staging as follows: stage 1 (infected 1–7 days), stage 2 (infected 8–14 days), and stage 3 (infected over 14 days). Furthermore, this system showed excellent sensor characteristics with a rapid response time of 15 min [11]. For case of electrochemical sensor, Vadlamani et al. 2020 designed an electrode composed of cobalt-functional TiO2 nanotubes (Co-TNT) as electrochemical sensor for the rapid detection of SARS-CoV-2 at low concentration range from 14–1400 nM with LOD of ~0.7 nM. The authors then used this system for real-time concentration measurements, showing a linear response in detecting viral proteins within concentration range for approximately 30 s in saliva and nasal secretions. The sensitivity of this approach can also be improved by using longer Co-TNTs due to higher surface area results in higher response rates, thus higher electric current can be obtained even at lower protein concentrations [12]. Furthermore, an advanced nanomaterial-based electrochemical biosensor to detect SARS-CoV-2 antibodies within seconds was successfully developed to enhance the rapid diagnosis of COVID-19 for the better treatment and prevention of diseases [13]. The three-dimensional (3D) electrodes were printed using 3D nanoprinting and were coated with nanoflakes of reduced-graphene-oxide (rGO); specific viral antigens were then immobilized on the rGO nanoflakes. The electrode was then integrated with a microfluidic device and applied as an electrical immunosensor. In the presence of antibodies against the SARS-CoV-2 S1 protein, the antibodies selectively bound to the antigens due to their strong immunoaffinity, leading to a change in impedance of the electrical circuit, which is detected via impedance spectroscopy. Antibodies to SARS-CoV-2 S1 protein and its receptor-binding domain were detected by a smartphone-based user interface within 10 s with a wide concentration range from 1 fM to 20 nM at LOD of 2.8 × 10−15 and 16.9 × 10−15 M, respectively (Figure 4). Seo et al. 2020 developed the field-effect transistor based biosensing device with the support of antibody functionalized graphene sheets to detect SARS-CoV-2 protein in human nasopharyngeal swab specimens. This platform helped improve nano-sensor performance in clinical samples for over 1 min without significantly altering the sensing capacity to detect SARS-CoV-2 without requiring sample pretreatment or labeling. This device showed good sensitivity for the detection of SARS-CoV-2 spike protein at the concentration of 1 fg/mL in phosphate-buffered saline and 100 fg/mL in clinical transport medium. Moreover, the device could detect SARS-CoV-2 in the culture medium with LOD of 1.6 × 10 pfu/mL and clinical samples with LOD of 2.42 × 102 copies/mL, respectively. This fully reversible modular sensing platform is a viable candidate for continuous clinical monitoring [14]. Overall, these biosensing platforms mentioned above exhibits high sensitivity, selectivity, and the rapid ability of monitoring SARS-CoV-2 for application in diagnosis of COVID-19. However, it highly requires the technical professions and instruments to conduct the sensing process that could ensure the accurate diagnosis. The important mission for researchers is to achieve proper stability, remove unwanted noise, and make the products commercially applicable after conducting a successful clinical trial.
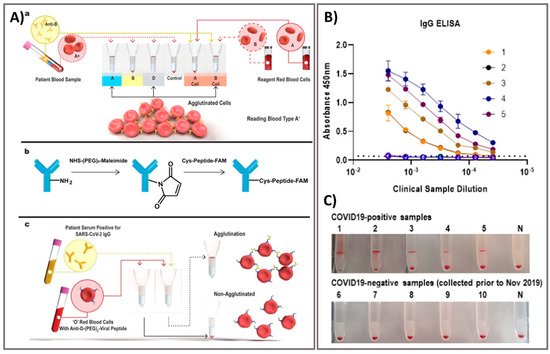
Figure 3. Rapid agglutination assays as serological testing for the detection of antibodies against SARS-CoV-2. (A) Schematic illustration of blood typing column agglutination test (CAT) with the brief antibody-peptide bioconjugates to produce the SARS-CoV-2 serological assay. (a) Pipette a mixture of reagent red blood cells (RRBCs) with patient samples onto a gel card containing separation media, followed by incubation of the card for 5–15 min. (b) The bioconjugation procedure to produce the antibody-peptide in two steps. (c) Antibody-peptide-coated RRBCs were incubated with a patient sample on a neutral gel prior to centrifugation to separate agglutinated RRBCs from free RRBCs for visual examination. Following optimization of the gel card assays to distinguish between SARS-CoV-2-positive samples and negative controls, 10 clinical samples were tested in both gel cards and by indirect IgG ELISA. (B) The results of indirect IgG ELISA comparing 10 samples, including PCR-confirmed SARS-CoV-2-positive samples and samples from healthy individuals collected before the SARS-CoV-2 outbreak. (C) Digital images of gel card assays recorded from experiments could identify positive/negative of antibodies, negative control noted (“N”). Reprinted with permission from [10]. ACS Sens. 2020, 5, 8, 2596–2603. Copyright 2020, American Chemical Society.
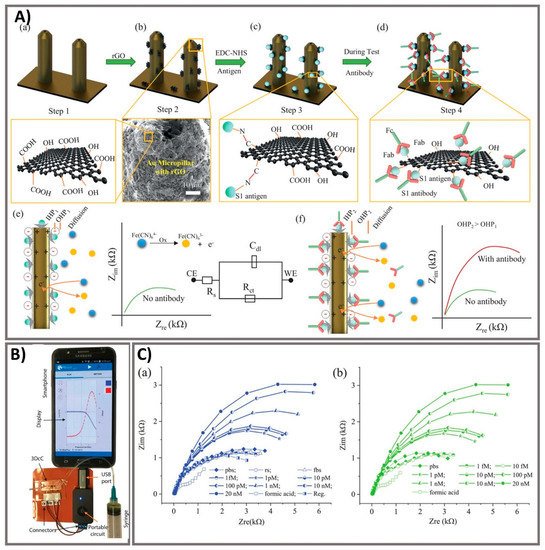
Figure 4. Ultra-rapid electrochemical immunosensor using aerosol jet nanoprinted reduced graphene oxide-coated 3D electrode for the detection of antibodies against SARS-CoV-2. (A) Functionalization of the 3D electrode and sensing operation. (a) AJ-printed gold micropillars prior to the surface treatment. (b) Coating rGO sheets onto the electrodes. (c) Immobilization of viral antigens onto rGO sheets. (d) Selective binding of antibody with specific antigens after introduction. (e) Schematics showing the sensing principle of the 3DcC device. (f) Schematic illustration of the Nyquist plot alternation via electrical impedance spectroscopy (EIS) before and after antibody introduction and binding with the antigens on the electrode surface. (B) The connection of the 3DcC device interfaced with a portable potentiostat to a smartphone via a USB-C connection for signal recording. (C) Sensing performance of antibodies against SARS-CoV-2 spike S1 antigen at different molar concentrations from 1 fM to 20 nM in (a) PBS solution and (b) after sensor regeneration using low-pH chemistry. Reprinted with permission from [13].
References
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555.
- Chow, F.W.-N.; Chan, T.T.-Y.; Tam, A.R.; Zhao, S.; Yao, W.; Fung, J.; Cheng, F.K.-K.; Lo, G.C.-S.; Chu, S.; Aw-Yong, K.L.; et al. A Rapid, Simple, Inexpensive, and Mobile Colorimetric Assay COVID-19-LAMP for Mass On-Site Screening of COVID-19. Int. J. Mol. Sci. 2020, 21, 5380.
- Thi, V.L.D.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Anders, S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12.
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282.
- Grant, B.D.; Anderson, C.E.; Williford, J.R. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309.
- Demey, B.; Daher, N.; Francois, C.; Lanoix, J.P.; Duverlie, G.; Castelain, S.; Brochot, E. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J. Infect. 2020, 81, e6–e10.
- Warren, A.D.; Kwong, G.A.; Wood, D.; Lin, K.Y.; Bhatia, S.N. Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proc. Natl. Acad. Sci. USA 2014, 111, 3671–3676.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277.
- Qin, Z.; Peng, R.; Baravik, I.K.; Liu, X. Fighting COVID-19: Integrated Micro- and Nanosystems for Viral Infection Diagnostics. Matter 2020, 3, 628–651.
- Alves, D.; Curvello, R.; Henderson, E. Rapid Gel Card Agglutination Assays for Serological Analysis Following SARS-CoV-2 Infection in Humans. ACS Sens. 2020, 5, 2596–2603.
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020, 92, 9454–9458.
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors 2020, 20, 5871.
- Ali, M.A.; Hu, C.; Jahan, S. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647.
- Seo, G.; Lee, G.; Kim, M.J. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142.
