Sugarcane (Saccharum spp.) is one of the most important industrial cash crops, contributing to the world sugar industry and biofuel production.
- Saccharum spp.
- biotic and abiotic stresses
- genetic engineering
1. Introduction
Sugarcane (Saccharum spp.) is an important agricultural crop for various subtropical and tropical countries, such as Brazil, India, Thailand, China, Australia, Pakistan, Philippines, Cuba, Colombia, and the USA [1]. It can produce various industrially valuable products, such as sugar, waxes, biofuels, and bio-fibres [2,3][2][3]. Globally, about 75% of sucrose is obtained from sugarcane, whereas the remaining 25% is obtained from Beta vulgaris [4]. There are two wild (S. robustum and S. spontaneum) and four cultivated (S. edule, S. barberi, S. sinense, and S. officinarum) main species in the Saccharum complex. However, the current commercial sugarcane cultivars, which are allopolyploids of high ploidy level, contain a narrow genetic range due to breeding via popular cultivars of the early 1900s, e.g., NCo310, Co419, and POJ2878 [5], whereas more recent cultivars have been developed via interspecific hybridisation of S. officinarum and S. spontaneum [6]. These commercial varieties are selected from an intervarietal hybrid population through clonal selection. The mechanism of diploidised meiosis makes them responsive to further breeding through hybridisation, even at the ploidy level of 10× or more. Gene introgression to produce intergeneric/interspecific hybrids with Erianthus, Miscanthus, Narenga, Sclerostachya, and Sorghum has been performed through intergeneric hybridisation. These sugarcane hybrids are fertile, vigorous, and could be maintained through vegetative propagation for various years; thus, they are termed perennial hybrids. Allopolyploid hybrids of Saccharum, due to their duplicated genomes, proved an ideal material for genetic modification research as compared to other seed-propagated annual plants. Transgenic protocols through Agrobacterium-mediated transformation and the biolistic approach have been used in sugarcane in the study of duplicated genes (mechanism of generating new genetic material during molecular evolution) [7]. Sugarcane can act as a model organism for studies on hybridisation, genome restructuring, chromosomal elimination, the effect of gene dosage, chromosomal bivalent pairing of higher polyploids, allelic variation, and so much more [8]. Erianthus arundinaceus presents a strong tolerance against abiotic stresses and could be widely used in modern breeding of sugarcane for producing cultivars with better stress tolerance, as well as enhanced sucrose content [9,10][9][10].
Considering the interest in rapidly improving sugarcane tolerance to stress under the current climate change scenario, conventional breeding programmes are time-consuming and ineffective [11]. Most importantly, several biotic and abiotic stresses can restrain the performance of sugarcane cultivars from emergence to harvesting [12]. Sugarcane’s heterogeneous nature, its vegetative propagation through stem cuttings, and poly-aneuploid genome make the research advances quite complex [13]. High sucrose/cellulose, ethanol-based biofuel production, and resistance to biotic and abiotic stresses are the distinct components considered by breeders for sugarcane improvement [14,15][14][15]. Beyond the overall improvement in carbohydrate contents, other traits such as higher emergence potential, plant vigour, and agronomic parameters—tillering, plant height, stem diameter, flowering, and resistance to biotic and abiotic stresses—are of paramount importance for achieving the highest potential of the sugarcane production system [16,17][16][17].
In the last decade, sugarcane genetic transformation and multiple omics technologies have received increasing attention, as they can open novel and unique research pathways [18]. The advent of recombinant DNA technology holds great potential, incorporating one or more specific genes via genetic engineering, to control single or multiple traits [19]. More recently, genome-editing techniques have been developed to achieve genetic modifications in which a gene/DNA fragment is replaced, deleted, or inserted into an organism genome using nucleases: meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and, at present, mostly the CRISPR/Cas9 system. During the process, the nucleases cause double-strand breaks (DSBs) at specific DNA regions; finally, non-homologous end joining (NHEJ) or homologous recombination (HR) result in the restoration of the induced DSBs [16]. Molecular markers associated with specific loci of the genome could be used for the analysis and detection of various genotypes in a gene pool of sugarcane [20]. Omics technologies, such as genomics, transcriptomics, proteomics, and metabolomics reveal complex connections between metabolites, proteins, and genes, which could help understand the genetic regulation and molecular mechanisms controlling both yield and stress resistance [21]. However, the limited use of this omics knowledge in sugarcane improvement against environmental stresses is presently frustrating. On the contrary, the development of transgenic sugarcane plants with significantly higher yield potential and better adaptability to fluctuating environmental conditions is moving towards a remarkable success [22]. Nevertheless, a comprehensive statistical genetic analysis is still required to identify genes associated with the increased yield and resistance to biotic and abiotic stresses of sugarcane cultivars.
2. Molecular Markers-Assisted Breeding (MAB)
2.1. Diverse Molecular Markers
The complex poly-aneuploid genome and narrow genetic base are the main limitations in breeding commercial varieties of sugarcane. Despite sugarcane nobilisation, little improvement has been achieved in enhancing sugar content [23]. Knowledge relevant to genetic diversity among elite cultivars is essential for further crop improvement and can be obtained through investigating morphological traits, pedigree analysis, or molecular markers [24]. Among these approaches, molecular markers have been widely used in the germplasm characterisation of Saccharum species. Besides, paternity analysis, phylogenetic relationship analysis, genetic mapping, QTL (quantitative trait loci) mapping, and MAS (molecular-assisted selection) have also received greater attention from researchers. Molecular markers and fragments/small regions/pieces of DNA sequence show polymorphism and reveal variations among different organisms (Figure 1). Different molecular markers, such as RAPDs (random amplified polymorphic DNA), RFLPs (restriction fragment length polymorphisms), AFLPs (amplified fragment length polymorphisms), SNPs (single nucleotide polymorphisms), TRAPs (target region amplification polymorphisms), SSRs (simple sequence repeats), and ISSRs (inter simple sequence repeats) have been widely used in the genetic study of sugarcane [20,25,26][20][25][26].
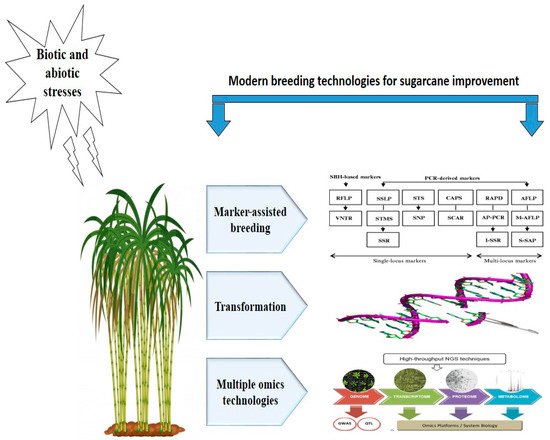
Figure 1. Modern breeding technologies for sugarcane genetic improvement against environmental stresses.
Genetic diversity in sugarcane germplasm was successfully identified using microsatellite DNA markers based on phenotypic traits and pedigree records [27]. Genetic diversity and phylogenetic relationship assessment by DNA markers can be crucial in germplasm classification and selecting breeding methods in sugarcane [28]. Molecular markers such as AFLP, SSR, and TRAPs have been successfully used in sugarcane to evaluate the genetic variations on a molecular level without the influence of environmental factors [29]. Genetic diversity analysis of 1002 accessions of sugarcane and related grasses, using 36 SSR markers, revealed the identification of 209 alleles associated with traits of agronomic importance [30]. Furthermore, 25 genotypes of sugarcane were also subjected to genetic diversity analysis through TRAP markers, which resulted in 70% genetic similarity between all accessions [31]. More recently, 150 of the most popular sugarcane accessions of China were successfully subjected to genetic diversity analysis with 21 fluorescence-labelled SSR markers and HPCE (high-performance capillary electrophoresis) for parental germplasm management [32]. Molecular markers help understand the genetic structures by identifying QTLs and developing genomic maps [33]. Incomplete genetic maps and linkages have been obtained using F1 individuals for Saccharum species such as S. officinarum [34], S. spontaneum [35], and a few modern sugarcane cultivars [36]. Genetic mapping of S. officinarum with 72 SSR and 40 AFLP primer pairs revealed 28 repulsion linkages [37]. A genetic mapping study of S. officinarum and S. spontaneum was carried out with AFLP, SRAP, and TRAP markers, which proved that AFLP is a better choice than SRAP and TRAP markers for sugarcane genetic mapping [24]. Whole-genome mapping of 183 sugarcane accessions was genotyped with 3327 DArT (diversity arrays technology), SSR, and AFLP markers to study 13 traits associated with morphology, the residual content of bagasse, resistance against diseases, and yield [38]. Phenotypic characterisation of 32 traits of 92 sugarcane varieties was performed using 174 SSR primers [39]. Linkage map construction and identification of genomic regions associated with traits of interest in the bi-parental population were carried out by QTL mapping [40].
2.2. Molecular Markers Related to Biotic Stresses
Genetic mapping of sugarcane is a challenge due to the presence of multiple alleles for a single gene. However, QTL mapping was proven to effectively discover various trait loci concerning yield [41] and disease resistance [42]. Several QTLs for disease resistance have been successfully identified in sugarcane, including 11 DNA markers for smut disease, five for resistance against fiji leaf gall, four for resistance against leaf scald and pachymetra root-rot [43], Bru1 for resistance against brown rust [44], and 18 QTLs for resistance against yellow leaf virus [45]. QTL mapping in association with AFLP and SSR markers resulted in the identification of one QTL link for resistance against yellow leaf spot disease [46]. Similarly, another QTL mapping via AFLP and SSR markers identified one major QTL link against the sugarcane yellow leaf virus [47]. Chilo sacchariphagus, the spotted-stem borer (SSB), is one of the major sugarcane pests. To understand its genetic basis, QTL mapping was performed with AFLP, RFLP, and SSR markers and resulted in identifying nine QTLs, ranging in phenotypic variation from 6 to 10% [48]. Additionally, genome regions associated with sugarcane resistance against leaf scald disease were successfully identified by QTL mapping with SNPs, SSRs, and ISSRs [49].
2.3. Molecular Markers Related to Abiotic Stresses
Molecular markers have been utilised to select resistant genotypes against abiotic stresses, including drought, salinity, or cold, in major crops such as wheat, rice, sugarcane, maize, sorghum, and barley [50]. For example, AFLP, RAPD, and SSR markers have been widely used for improving the adaptability of sugarcane to abiotic stresses. Thus, RAPD markers analysis resulted in identifying molecular markers associated with resistance to drought and salinity [51,52][51][52]. Drought tolerance screening of 23 sugarcane genotypes was carried out using a SCAR marker generated from OPAK-12724 (RAPD DNA-sequence). This study identified 12 drought-tolerant genotypes, indicating that SCAR was 92% specific in the selection and occupied the same locus as OPAK-12724 [53]. Molecular selection in eight sugarcane genotypes (BOT-41, Co-775, Co-997, F-153, F-161, GT54-9, G84-47, and Sp80-32-80) regarding drought tolerance was also performed using RAPD, ISSR, and R-ISSR (combined RAPD and ISSR) markers. Among these, the use of R-ISSR was proved to be more efficient than the independent selection with RAPD or ISSR [54]. A further selection of these genotypes was carried out with AFLP primers by amplifying 886 amplicons with 6.2% polymorphism; this analysis established Co-997 as the most tolerant and Co-775 as the most susceptible to drought of the eight tested genotypes [55].
Sugarcane, being a glycophyte, exhibits growth reduction, nutritional imbalance, low-sprout emergence, low sugar content, and low productivity under salt stress [56]. Molecular markers can be used to track genetic loci related to salt resistance, avoiding time-consuming phenotypic measurements. Indeed, several PCR-based markers have been employed to assess the genetic diversity of sugarcane cultivars regarding their relative salt tolerance. For example, in vitro mutagenesis and RAPD markers were used for the selection and molecular characterisation of salt-tolerant lines in S. officinarum; RAPD markers indicated the genetic polymorphism in the control and salt-tolerant lines [57]. Screening of salt-tolerant and susceptible lines in sugarcane was also conducted with 15 ISSR markers, based on the similarity index among cultivars. This study proved that the ISSR markers effectively assessed the genetic diversity of sugarcane cultivars [58]. Characterisation between parents and mutant lines (drought and salt-tolerant) was carried out with RAPD markers under tissue culture selection pressure. Sugarcane embryonic calli treated with ethyl-methane sulphonate (EMS) were grown under drought and salt stress conditions, and tolerant lines were separated from the controls, based on the polymorphisms revealed by the RAPD profiles [59]. Similarly, in vitro molecular profiling of sugarcane variants was carried out using RAPD markers under water deficit and salt stress treatments [60]. Furthermore, molecular characterisation of 18 sugarcane genotypes under salt stress was undertaken using five TRAP markers. A similarity coefficient of 0.72 was observed, with 25 to 100% polymorphisms, which revealed a limited variation between the tested genotypes under salt stress [61]. Overall, molecular markers are powerful tools for identifying phenotypic and genetic diversity and provide a platform for introducing new desirable traits in sugarcane against abiotic and biotic stresses.
References
- Sucden. Available online: (accessed on 13 January 2021).
- Singh, A.; Lal, U.R.; Mukhtar, H.M.; Singh, P.S.; Shah, G.; Dhawan, R.K. Phytochemical profile of sugarcane and its potential health aspects. Phcog. Rev. 2015, 9, 45.
- Iryani, D.A.; Kumagai, S.; Nonaka, M.; Sasaki, K.; Hirajima, T. Characterization and production of solid biofuel from sugarcane bagasse by hydrothermal carbonization. Waste Biomass Valori. 2017, 8, 1941–1951.
- Kroger, M.; Meister, K.; Kava, R. Low—Calorie sweeteners and other sugar substitutes: A review of the safety issues. Food Sci. 2006, 5, 35–47.
- Hunsigi, G. Production of Sugarcane Theory and Practice; Springer: Berlin, Germany; Heidelberg, Germany, 1993; pp. 12–28.
- Fukuhara, S.; Terajima, Y.; Irei, S.; Sakaigaichi, T.; Ujihara, K.; Sugimoto, A.; Matsuoka, M. Identification and characterization of intergeneric hybrid of commercial sugarcane (Saccharum spp. hybrid) and Erianthus arundinaceus (Retz.) Jeswiet. Euphytica 2013, 189, 321–327.
- Arvinth, S.; Arun, S.; Selvakesavan, R.K.; Srikanth, J.; Mukunthan, N.; Kumar, P.A.; Premachandran, M.N.; Subramonian, N. Genetic transformation and pyramiding of aprotinin-expressing sugarcane with cry1Ab for shoot borer (Chilo infuscatellus) resistance. Plant Cell Rep. 2010, 29, 383–395.
- Premachandran, M.N.; Prathima, P.T.; Lekshmi, M. Sugarcane and polyploidy: A review. J. Sugarcane Res. 2011, 1, 1–15.
- Yan, J.; Zhang, J.; Sun, K.; Chang, D.; Bai, S.; Shen, Y.; Huang, L.; Zhang, J.; Zhang, Y.; Dong, Y. Ploidy level and DNA content of Erianthus arundinaceus as determined by flow cytometry and the association with biological characteristics. PLoS ONE 2016, 11, e0151948.
- Cai, Q.; Aitken, K.S.; Fan, Y.H.; Piperidis, G.; Liu, X.L.; McIntyre, C.L.; Huang, X.Q.; Jackson, P. Assessment of the genetic diversity in a collection of Erianthus arundinaceus. Genet. Resour. Crop Evol. 2012, 59, 1483–1491.
- Scortecci, K.C.; Creste, S.; Calsa, J.T.; Xavier, M.A.; Landell, M.G.; Figueira, A.; Benedito, V.A. Challenges, opportunities and recent advances in sugarcane breeding. Plant Breed 2012, 1, 267–296.
- Meena, M.R.; Kumar, R.; Chinnaswamy, A.; Karuppaiyan, R.; Kulshreshtha, N.; Ram, B. Current breeding and genomic approaches to enhance the cane and sugar productivity under abiotic stress conditions. 3 Biotech 2020, 10, 440.
- Tew, T.L.; Cobill, R.M. Genetic improvement of sugarcane (Saccharum spp.) as an energy crop. In Genetic Improvement of Bioenergy Crops; Springer: New York, NY, USA, 2008; pp. 273–294.
- Tahir, M.; Rahman, H.; Gul, R.; Ali, A.; Khalid, M. Genetic divergence in sugarcane genotypes. J. Exp. Agric. Int. 2013, 3, 102–109.
- Singh, S.P.; Singh, P.; Sharma, B.L. Methods to improve germination in sugarcane. In Emerging Trends of Plant Physiology for Sustainable Crop Production; CRC Press: Washington, DC, USA, 2018; pp. 331–344.
- Güell, M.; Yang, L.; Church, G.M. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinform 2014, 30, 2968–2970.
- Sanghera, G.S.; Malhotra, P.K.; Singh, H.; Bhatt, R. Climate change impact in sugarcane agriculture and mitigation strategies. In Harnessing Plant Biotechnology and Physiology to Stimulate Agricultural Growth; The Journal of Plant Science and Research: New Delhi, India, 2019; pp. 99–115.
- Khan, M.T.; Khan, I.A.; Yasmeen, S. Genetically modified sugarcane for biofuels production: Status and perspectives of conventional transgenic approaches, RNA interference, and genome editing for improving sugarcane for biofuels. In Sugarcane Biofuels; Springe: Cham, Switzerland, 2019; pp. 67–96.
- Wright, S. Recombinant DNA technology and its social transformation. Osiris 1986, 1, 303–360.
- Ali, A.; Pan, Y.B.; Wang, Q.N.; Wang, J.D.; Chen, J.L.; Gao, S.J. Genetic diversity and population structure analysis of Saccharum and Erianthus genera using microsatellite (SSR) markers. Sci. Rep. 2019, 9, 1–10.
- Devarumath, R.M.; Kalwade, S.B.; Kulkarni, P.A.; Sheelavanthmath, S.S.; Suprasanna, P. Integrating OMICS Approaches in Sugarcane Improvement. OMICS Applications in Crop Science; CRC Press: New York, NY, USA, 2013; pp. 191–250.
- Mirajkar, S.J.; Devarumath, R.M.; Nikam, A.A.; Sushir, K.V.; Babu, H.; Suprasanna, P. Sugarcane (Saccharum spp.): Breeding and genomics. In Advances in Plant Breeding Strategies: Industrial and Food Crops; Springer: Cham, Switzerland, 2019; pp. 363–406.
- Liu, P.; Chandra, A.; Que, Y.; Chen, P.H.; Grisham, M.P.; White, W.H.; Dalley, C.D.; Tew, T.L.; Pan, Y.-B. Identification of quantitative trait loci controlling sucrose content based on an enriched genetic linkage map of sugarcane (Saccharum spp. hybrids) cultivar ‘LCP 85–384’. Euphytica 2016, 207, 527–549.
- Alwala, S.; Kimbeng, C.A.; Veremis, J.C.; Gravois, K.A. Linkage mapping and genome analysis in Saccharum interspecific cross using AFLP, SRAP and TRAP markers. Euphytica 2008, 164, 37–51.
- Palhares, A.C.; Rodrigues-Morais, T.B.; Van Sluys, M.A.; Domingues, D.S.; Maccheroni, W.; Jordão, H.; Souza, A.P.; Marconi, T.G.; Mollinari, M.; Gazaffi, R.; et al. A novel linkage map of sugarcane with evidence for clustering of retrotransposon-based markers. BMC Genet. 2012, 13, 51.
- Manimekalai, R.; Suresh, G.; Govinda Kurup, H.; Athiappan, S.; Kandalam, M. Role of NGS and SNP genotyping methods in sugarcane improvement programs. Critic. Rev. Biotechnol. 2020, 40, 1–16.
- Cordeiro, G.M.; Pan, Y.B.; Henry, R.J. Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Sci. 2003, 165, 181–189.
- You, Q.; Pan, Y.B.; Xu, L.P.; Gao, S.W.; Wang, Q.N.; Su, Y.C.; Yang, Y.Q.; Wu, Q.B.; Zhou, D.G.; Que, Y.X. Genetic diversity analysis of sugarcane germplasm based on fluorescence—Labeled simple sequence repeat markers and a capillary electrophoresis—Based genotyping platform. Sugar Tech. 2016, 18, 380–390.
- Creste, S.; Sansoli, D.M.; Tardiani, A.C.S.; Silva, D.N.; Goncalves, F.K.; Favero, T.M.; Medeiros, C.N.F.; Festucci, C.S.; Carlini-Garcia, L.A.; Landell, M.G.; et al. Comparison of AFLP, TRAP and SSRs in the estimation of genetic relationships in sugarcane. Sugar Tech. 2010, 12, 150–154.
- Nayak, S.N.; Song, J.; Villa, A.; Pathak, B.; Ayala-Silva, T.; Yang, X.; Todd, J.; Glynn, N.C.; Kuhn, D.N.; Glaz, B.; et al. Promoting utilization of Saccharum spp. genetic resources through genetic diversity analysis and core collection construction. PLoS ONE 2014, 9, e110856.
- Singh, R.B.; Singh, B.; Singh, R.K. Study of genetic diversity of sugarcane (Saccharum) species and commercial varieties through TRAP molecular markers. Indian J. Plant Physiol. 2017, 22, 332–338.
- Wu, J.; Wang, Q.; Xie, J.; Pan, Y.B.; Zhou, F.; Guo, Y.; Chang, H.; Xu, H.; Zhang, W.; Zhang, C.; et al. SSR marker-assisted management of parental germplasm in sugarcane (Saccharum spp. hybrids) breeding programs. Agron 2019, 9, 449.
- Pinto, L.R.; Garcia, A.A.F.; Pastina, M.M.; Teixeira, L.H.M.; Bressiani, J.A.; Ulian, E.C.; Bidoia, M.A.P.; Souza, A.P. Analysis of genomic and functional RFLP derived markers associated with sucrose content, fiber and yield QTLs in a sugarcane (Saccharum spp.) commercial cross. Euphytica 2010, 172, 313–327.
- Guimaraes, C.T.; Sills, G.R.; Sobral, B.W.S. Comparative mapping of Andropogoneae: Saccharum L. (sugarcane) and its relation to sorghum and maize. Proc. Natl. Acad. Sci. USA 1997, 94, 14262–14266.
- Silva, J.A.G.; Sorrells, M.E.; Burnquist, W.; Tanksley, S.D. RFLP linkage map and genome analysis of Saccharum spontaneum. Genome 1993, 36, 782–791.
- Zhou, J.R.; Sun, H.D.; Ali, A.; Rott, P.C.; Javed, T.; Fu, H.Y.; Gao, S.J. Quantitative proteomic analysis of the sugarcane defense responses incited by Acidovorax avenae subsp. avenae causing red stripe. Ind. Crops Prod. 2021, 162, 113275.
- Aitken, K.S.; Jackson, P.A.; McIntyre, C.L. QTL identified for sugar related traits in a sugarcane (Saccharum spp.) cultivar 9 S. officinarum population. Theor. Appl. Genet. 2007, 112, 1306–1317.
- Gouy, M.; Rousselle, Y.; Chane, A.T.; Anglade, A.; Royaert, S.; Nibouche, S.; Costet, L. Genome wide association mapping of agro—Morphological and disease resistance traits in sugarcane. Euphytica 2015, 202, 269–284.
- Siraree, A.; Banerjee, N.; Kumar, S.; Khan, M.S.; Singh, P.K.; Sharma, S.; Singh, R.K.; Singh, J. Identification of marker—Trait associations for morphological descriptors and yield component traits in sugarcane. Physiol. Mol. Biol. Plants 2017, 23, 185–196.
- Collard, B.C.; Jahufer, M.Z.; Brouwer, J.B.; Pang, E.C. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196.
- Singh, R.K.; Singh, S.P.; Tiwari, D.K.; Srivastava, S.; Singh, S.B.; Sharma, M.L.; Singh, R.; Mohapatra, T.; Singh, N.K. Genetic mapping and QTL analysis for sugar yield-related traits in sugarcane. Euphytica 2013, 191, 333–353.
- Aitken, K.S. Introgression of a large effect QTL for smut resistance inherited from S. Spontaneum. In Proceedings of the Plant and Animal Genome XXIV Conference, San Diego, CA, USA, 8–13 January 2016.
- Wei, X.; Jackson, P.A.; McIntyre, C.L.; Aitken, K.S.; Croft, B. Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor. Appl. Genet. 2006, 114, 155–164.
- Daugrois, J.H.; Grivet, L.; Roques, D.; Hoarau, J.Y.; Lombard, H.; Glaszmann, J.C.; D’Hont, A.A. A putative major gene for rust resistance linked with a RFLP marker in sugarcane cultivar ‘R570’. Theor. Appl. Genet. 1996, 92, 1059–1064.
- You, Q.; Yang, X.; Peng, Z.; Islam, M.S.; Sood, S.; Luo, Z.; Comstock, J.; Xu, L.; Wang, J. Development of an Axiom Sugarcane100K SNP array for genetic map construction and QTL identification. Theor. Appl. Genet. 2019, 132, 2829–2845.
- Aljanabi, S.M.; Parmessur, Y.; Kross, H.; Dhayan, S.; Saumtally, S.; Ramdoyal, K.; Autrey, L.J.; Dookun-Saumtally, A. Identification of a major quantitative trait locus (QTL) for yellow spot (Mycovellosiella koepkei) disease resistance in sugarcane. Mol. Breed. 2007, 19, 1–14.
- Costet, L.; Raboin, L.; Payet, M.; D’Hont, A.; Nibouche, S. A major quantitative trait allele for resistance to the Sugarcane yellow leaf virus (Luteoviridae). Plant Breed. 2012, 131, 637–640.
- Nibouche, S.; Raboin, L.M.; Hoarau, J.Y.; D’Hont, A.; Costet, L. Quantitative trait loci for sugarcane resistance to the spotted stem borer Chilo sacchariphagus. Mol. Breed. 2012, 29, 129–135.
- Gutierrez, A.; Hoy, J.; Kimbeng, C.; Baisakh, N. Identification of genomic regions controlling leaf scald resistance in sugarcane using a bi—Parental mapping population and selective genotyping by sequencing. Front. Plant Sci. 2018, 9, 877.
- Winicov, I. Molecular markers and abiotic stresses. In Molecular Techniques in Crop Improvement; Springer: New York, NY, USA, 2010.
- Abdel-Tawab, F.M.; Fahmy, E.M.; Allam, A.I.; El-Rashidy, H.A.; Shoaib, R.M. Development of RAPD and SSR marker associated with stress tolerance and some technological traits and transient transformation of sugarcane (Saccharum spp.). In Proceedings of the International Confernence. The Arab Region and Africa in the World Sugar Context, Aswan, Egypt, 9–12 March 2003; pp. 10–12.
- Abdel-Tawab, F.M.; Fahmy, E.M.; Allam, A.I.; El-Rashidy, H.A.; Shoaib, R.M. Marker assisted selection for abiotic stress tolerance in sugarcane (Saccharum spp.). Egypt. J. Agric. Res. 2003, 81, 635–646.
- Srivastava, M.K.; Li, C.N.; Li, Y.R. Development of sequence characterized amplified region (SCAR) marker for identifying drought tolerant sugarcane genotypes. Aus. J. Crop Sci. 2012, 6, 763.
- Khaled, K.; El-Sherbeny, S.; Abdelhadi, A. R-ISSR as marker assisted selection for drought tolerance in sugarcane. J. Agric. Chem. Biotechnol. 2017, 8, 91–97.
- Khaled, K.A.; El-Arabi, N.I.; Sabry, N.M.; El-Sherbiny, S. Sugarcane genotypes assessment under drought condition using amplified fragment length polymorphism. Biotechnology 2018, 17, 120–127.
- Chaum, S.; Chuencharoenm, S.; Mongkolsiriwatanam, C.; Ashrafm, M.; Kirdmanee, C. Screening sugarcane (Saccharum sp.) genotypes for salt tolerance using multivariate cluster analysis. Plant Cell Tiss. Org. Cult. 2012, 110, 23–33.
- Suprasanna, P.; Patade, V.Y.; Vaidya, E.R.; Patil, V.D. Radiation induced in vitro mutagenesis, selection for salt tolerance and characterization in sugarcane. In Induced Plant Mutations in the Genomics Era: Proceedings of an International Joint FAO/IAEA Symposium; International Atomic Energy Agency: Vienna, Austria, 2009; pp. 145–147.
- Markad, N.R.; Kale, A.A.; Pawar, B.D.; Jadhav, A.S.; Patil, S.C. Molecular characterization of sugarcane (Saccharum officinarum L.) genotypes in relation to salt tolerance. Bioscan 2014, 9, 1785–1788.
- Gadakh, S.; Patel, D.; Singh, D. Use of RAPD markers to characterize salt and drought lines of sugarcane. Int. J. Adv. Res. Biol. Sci. 2017, 4, 50–57.
- Yadav, P.V.; Suprasanna, P.; Gopalrao, K.U.; Anant, B.V. Molecular profiling using RAPD technique of salt and drought tolerant regenerants of sugarcane. Sugar Tech. 2006, 8, 63–68.
- Farsangi, F.J.; Thorat, A.S.; Devarumath, R.M. Molecular characterization of sugarcane genotypes for their salinity and susceptibility using TRAP markers. Int. J. Curr. Res. 2018, 10, 68947–68951.
