The COVID-19 pandemic represents an unprecedented opportunity to exploit the advantages of personalized medicine for the prevention, diagnosis, treatment, surveillance and management of a new challenge in public health. COVID-19 infection is highly variable, ranging from asymptomatic infections to severe, life-threatening manifestations. Personalized medicine can play a key role in elucidating individual susceptibility to the infection as well as inter-individual variability in clinical course, prognosis and response to treatment. Integrating personalized medicine into clinical practice can also transform health care by enabling the design of preventive and therapeutic strategies tailored to individual profiles, improving the detection of outbreaks or defining transmission patterns at an increasingly local level. SARS-CoV2 genome sequencing, together with the assessment of specific patient genetic variants, will support clinical decision-makers and ultimately better ways to fight this disease. Additionally, it would facilitate a better stratification and selection of patients for clinical trials, thus increasing the likelihood of obtaining positive results. Lastly, defining a national strategy to implement in clinical practice all available tools of personalized medicine in COVID-19 could be challenging but linked to a positive transformation of the health care system.
- personalized medicine
- precision medicine
- Covid-19
- SARS CoV2
- epidemiology
- host genetics
- viral genome
1. Introduction
2. Translating Personalized Medicine into Clinical Practice: The Andalusian Experience
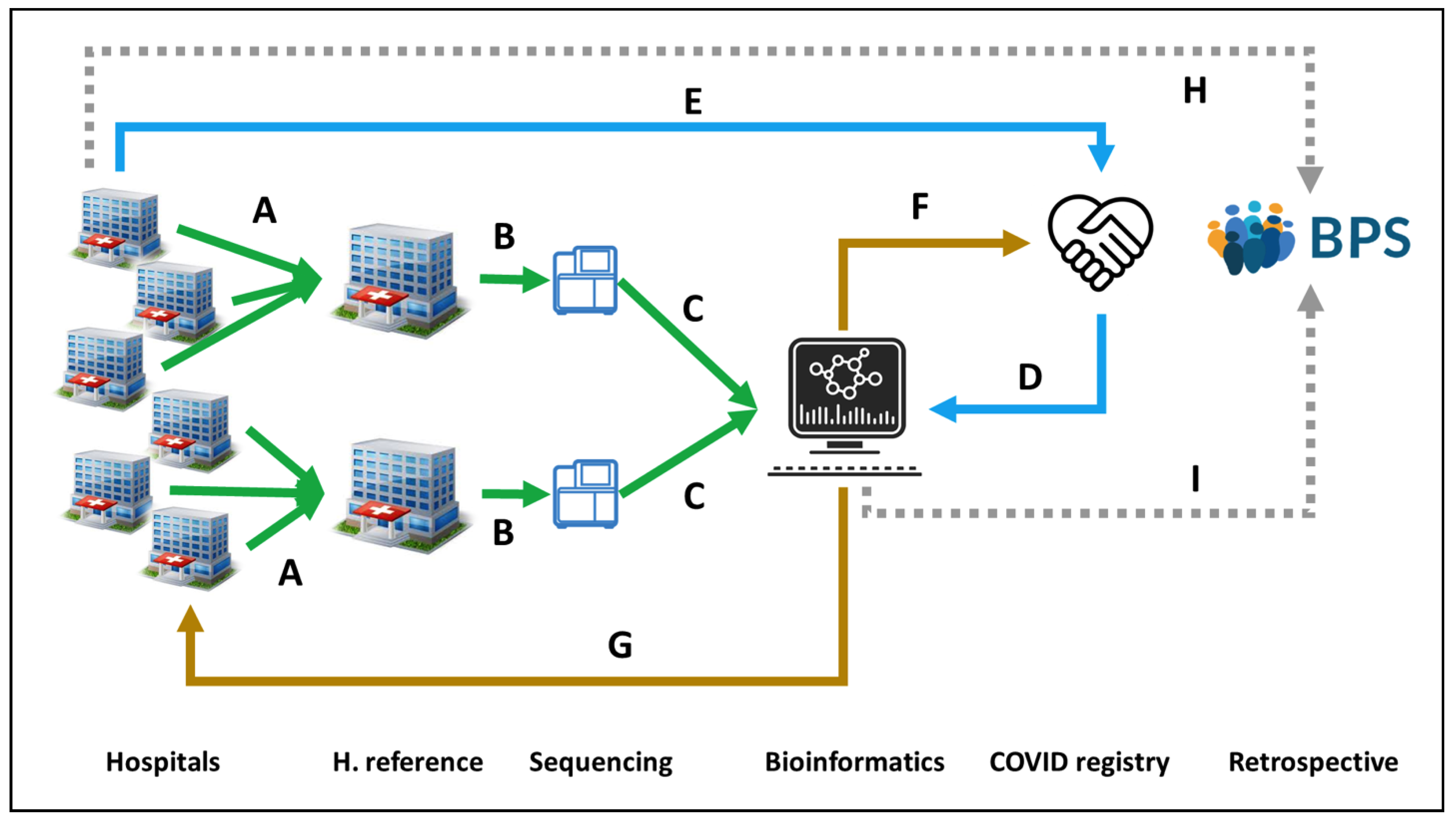 Figure 1. Circuit for COVID19 genomic surveillance. (A) Two reference Hospitals, San Cecilio and Virgen del Rocio, collect SARS-CoV-2 samples from Eastern and Western Andalusia, respectively. (B) Samples are sent to GENYO or IBIS sequencing facilities. (C) Sequencing data are sent to the Bioinformatics Area for processing and (D) linking to metadata from the COVID registry, (E) previously collected from the Hospitals. (F) Bioinformatics reports relevant epidemiological information to the COVID registry and (G) information on lineages and variants to the Hospitals for supporting clinical decisions. (H) Clinical data on COVID-19 patients recorded in the Hospitals is stored in the BPS. (I) Viral genomes are also stored in BPS linked to the rest of the patient’s clinical data for further secondary use.
Figure 1. Circuit for COVID19 genomic surveillance. (A) Two reference Hospitals, San Cecilio and Virgen del Rocio, collect SARS-CoV-2 samples from Eastern and Western Andalusia, respectively. (B) Samples are sent to GENYO or IBIS sequencing facilities. (C) Sequencing data are sent to the Bioinformatics Area for processing and (D) linking to metadata from the COVID registry, (E) previously collected from the Hospitals. (F) Bioinformatics reports relevant epidemiological information to the COVID registry and (G) information on lineages and variants to the Hospitals for supporting clinical decisions. (H) Clinical data on COVID-19 patients recorded in the Hospitals is stored in the BPS. (I) Viral genomes are also stored in BPS linked to the rest of the patient’s clinical data for further secondary use.3. Concluding Remarks
References
- Loucera, C.; Esteban-Medina, M.; Rian, K.; Falco, M.M.; Dopazo, J.; Peña-Chilet, M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal Transduct. Target. Ther. 2020, 5, 290.
- Friedman, J.M.; Jones, K.L.; Carey, J.C. Exome Sequencing as Part of a Multidisciplinary Approach to Diagnosis-Reply. JAMA 2020, 324, 2445–2446.
- Muñoyerro-Muñiz, D.; Goicoechea-Salazar, J.; García-León, F.; Laguna-Tellez, A.; Larrocha-Mata, D.; Cardero-Rivas, M. Health record linkage: Andalusian health population database. Gaceta Sanitaria 2019, 34, 105–113.
- BPS and Research. Andalusian Health Population Database (Base Poblacional de Salud), 2020. Available online: (accessed on 3 January 2021).
- García-León, F.; Villegas-Portero, R.; Goicoechea-Salazar, J.; Muñoyerro-Muñiz, D.; Dopazo, J. Impact assessment on data protection in research projects. Gaceta Sanitaria 2020, 34, 521–523.
- Clinical Bioinformatics Area. Progress and Health Foundation, 2017. Available online: (accessed on 3 April 2021).
