Freshwater ecosystems and their associated biota have been negatively impacted by the human development of water resources. Fundamental to restoration activities for target species is an understanding of the factors affecting population decline or recovery. Within Australia’s Murray–Darling Basin, recovery efforts to address the population decline of native freshwater fish include stock enhancement, habitat restoration, and the delivery of environmental water. Essential to guiding future management actions is information to assess the efficacy of these efforts. We undertook a study to investigate whether natural spawning and recruitment, stock enhancement, or a combination of the two is contributing to sustaining populations of golden perch (
Macquaria ambigua
) in the highly regulated Lachlan River, Australia. Otolith microchemistry and genetic analyses were used as complementary tools to determine the source (hatchery origin or wild-spawned) of existing populations in the catchment. We identified that natural spawning and recruitment was contributing to riverine populations in some years but that populations were heavily reliant on stocking. It was not possible to distinguish hatchery and wild-born fish using genetic tools, highlighting the value of using multiple lines of evidence to establish causal mechanisms contributing to population recovery.
- isheries management
- conservation ecology
- stocking
- otolith
- freshwater fish
- Murray–Darling Basin
Otolith Micro-Chemical Composition
Four of the nine elements analyzed showed a significant difference in concentration (ug·g
) between the otolith core and edge for wild-caught fish from the Lachlan River (
Mg,
Sr,
Mn, and
Zn; Student’s
-test
-values all < 0.01) (Figure 1, panel LL_edge vs. LL_core and UL_edge vs. UL_core).
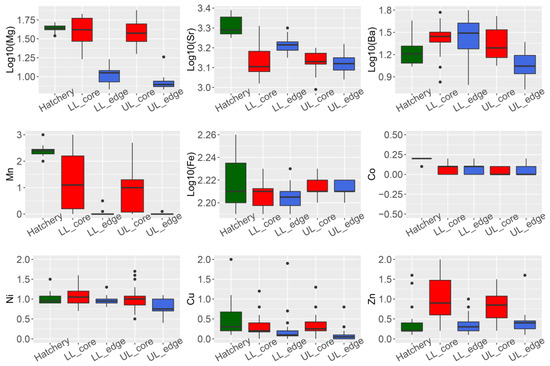
Summary of trace element composition (ug·g
) in otoliths of
(golden perch) from three localities. Hatchery = hatchery sourced YOY samples (n = 19). LL = samples from the lower Lachlan (downstream Lake Brewster) (n = 24); UL = samples from the upper Lachlan (upstream Wyangala Dam) (n = 20); core = measurements from the core of the otolith; edge = measurements from the edge of the otolith.
The NMDS clearly separated the chemical signatures from the edges of otoliths from fish from the Lachlan River and those of the hatchery (YOY) samples (stress: 0.07) (Figure 2a). A PERMANOVA test with 999 bootstrapping using the Bray–Curtis dissimilarity index found a significant difference between the clusters (
-value = 0.001,
model = 76.35, ANOVA for homogeneity of multivariate dispersion
= 0.39), indicating a unique chemical signature for each locality. The analyses were then repeated on the lower Lachlan and upper Lachlan edge and core samples separately (Figure 2b,c). For the lower Lachlan, this revealed two distinct groups comprising core and edge samples (Figure 2b). The signature from the hatchery reference samples was more similar to the lower Lachlan core chemical signature than the edge signature of the lower Lachlan River individuals. In addition, a small group of individuals displayed similar core and edge signatures (Figure 2b), suggesting wild-born individuals from the lower Lachlan. Cluster analysis using elements significantly contributing to the core and edge variation (
Mg,
Sr,
Mn, and
Zn) identified three primary groups (clusters) representing predominantly edge, core, and hatchery signatures. This analysis clearly identified six
individuals for which core signatures were clustered with the edge signature indicating natural recruitment in the lower Lachlan catchment. Within the core and hatchery (YOY) groups, four individuals displayed core signatures that clustered with the hatchery reference group while the remaining individuals displayed core signatures that grouped separately (Figure 2d).
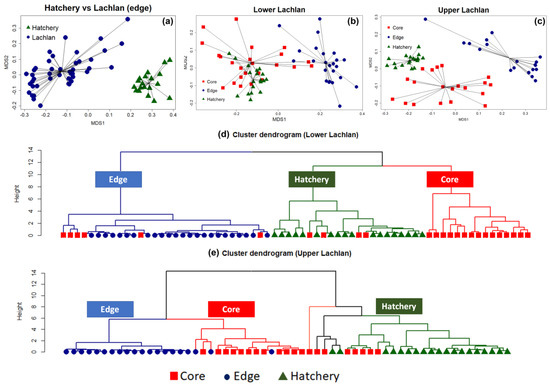
Non-metric multidimensional scaling (NMDS) and cluster analysis showing the presence of both wild-born and hatchery-bred stocked
from the Lachlan River. (
) NMDS spider plot between the edge of the Lachlan river sample and the hatchery reference individuals (Hatchery YOY); (
) NMDS spider plot among the edge and core of the lower Lachlan river sample with the presence of the hatchery reference; (
) NMDS spider plot among the edge and core of the upper Lachlan river sample with the presence of the control group (Hatchery YOY); (
) Cluster dendrogram of the lower Lachlan samples and the hatchery reference based on a Euclidean distance matrix representing the presence of natural recruitment in the lower Lachlan catchment; (
) Cluster dendrogram of the upper Lachlan samples and hatchery reference group based on a Euclidean distance matrix representing no natural recruitment with two distinct clusters between the Lachlan River samples and hatchery reference group.
In the upper Lachlan River samples, the NMDS revealed three distinct groups comprising core, edge, and hatchery reference samples, but the cluster analysis using only those elements significantly contributing to the core and edge variation suggests more complex groupings within the data set (Figure 2e). The cluster analysis identified two primary clusters separating the core and edge measurements of the Lachlan River samples from the hatchery reference collections. Five tentative sub-clusters belonging to two major groups of Lachlan River samples and hatchery samples were evident; however, the groupings were less distinct within the clusters (Figure 2e). The results suggest none of the upper Lachlan samples were wild-born, while five individuals displayed a core signature that clustered with the hatchery reference group.
Our analysis revealed the presence of both hatchery-bred stocked and wild-born
from the Lachlan River. Among the 44 fish collected from Lachlan River, a total of six
were identified as wild-born fish (14%), while the rest of them were either hatchery-bred stocked fish or from unknown sources (Figure 3). The observation of annual rings in the otolith revealed that all six wild-born
sampled from the lower Lachlan, were spawned in 2002, 2007, 2009, 2014 (one fish each year), and 2010 (two fish). A comparison between the daily discharge data from the lower Lachlan (Figure 3) revealed no clear association between high flow events and the spawning year of the six wild-born fish.
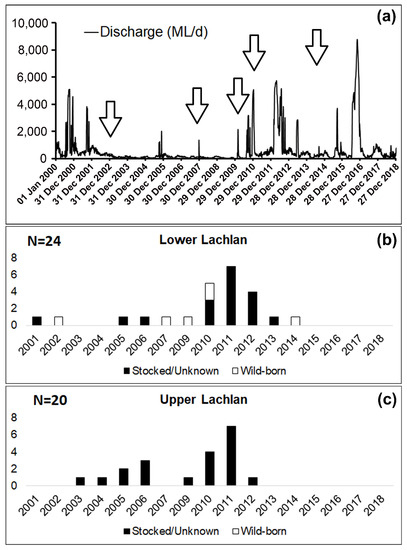
Daily discharge from the lower Lachlan since 2000 (Source: WaterNSW, http://realtimedate.waternsw.com.au/), and the number of stocked/unknown and wild-born fish from the Lachlan River and their spawning year. (
) Daily discharge (ML/day) at Hillston Weir (lat: −33.4873 lon:145.504) from 1 January 2000 to 31 December 2018; (
) Samples from the lower Lachlan (N = 24); (
) Samples from the upper Lachlan (N = 20). White bars represent the number of wild-born fish, and the black bars represent the number of fish either stocked or from unknown sources.
Genetic Data Analysis
The Diversity Arrays Technology (DArT) pipeline scored 5207 single nucleotide polymorphisms (SNPs) and 6956 SilicoDArT (presence-absence) markers after filtering based on polymorphic information content (>0.025). For the present study, we filtered out 2124 SNP markers based on the call ratio (0.90) and reproducibility average (0.90), resulting in a total of 3083 SNPs (null allele = 2.1%). Further filtering based on the Hardy–Weinberg equilibrium (HWE) was performed to remove markers under selection. Finally, 1481 SNPs were used for the present study. The same criteria (call ratio and polymorphic information content) were also used to filter out low-quality SilicoDArT markers. After filtering, 2902 (nNull allele = 5.7%) SilicoDArT markers were used for further analysis. Filtering for SilicoDArT loci under selection was not performed, as the allelic form of the loci is unknown (presence and absence of restriction fragment).
Despite a small cluster of hatchery reference individuals (hatchery YOY) (Figure 4a), the principal component analysis (PCA) suggests no specific clustering using SNP markers. High similarity among the Lachlan River samples and hatchery YOY and a lack of genetic structure means that the SNP markers could not conclusively distinguish the wild-born fish from the hatchery-bred stocked or unknown-source fish. We also performed a principal component analysis using only Lachlan River samples testing for differences between wild-born fish identified a priori and the rest from the otolith data (Figure 4b) and could find no genetic differences. The principal component analysis using SilicoDArT markers found a similar result, although it did indicate differences between the lower Lachlan fish (widely spread across PC1/Dim1 in the PCA plot) and upper Lachlan fish (a single cluster, which included the hatchery-bred YOY fish) (Figure 4c,d).
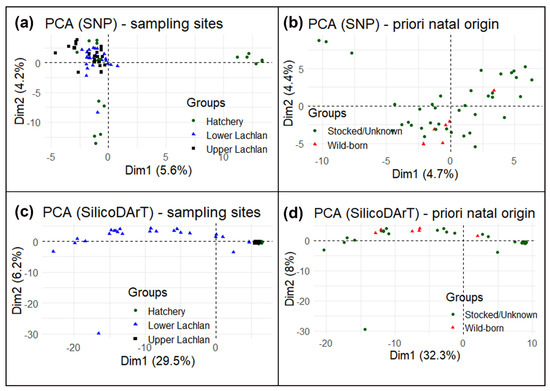
Principal component analysis (PCA) of
; (
) PCA plot using Single Nucleotide Polymorphism markers—plotting based on geographic locations such as lower Lachlan, Upper Lachlan, and Hatchery; (
) PCA plot of only Lachlan samples using Single Nucleotide Polymorphism markers—plotting based on natal origins such as wild-born fish and hatchery-bred stocked/unknown-source fish; (
) PCA plot using SilicoDArT markers–plotting based on geographic locations such as Lower Lachlan, Upper Lachlan, and Hatchery; (
) PCA plot of only Lachlan samples using SilicoDArT markers—plotting based on natal origins such as wild-born fish and hatchery-bred stocked/unknown-source fish.
Discussion
The aim of this study was to investigate whether there was evidence of the natural recruitment of the existing
population at the Lachlan River, or whether all the fish were derived from hatchery stocking. In doing so, we tested the effectiveness of genetic and otolith data as comparative tools. The analysis of otolith microchemistry identified limited natural recruitment along with the co-existence of wild-born and hatchery-bred stocked fish in the lower reach of the Lachlan River. While the data set was less clear about the origin of the upper Lachlan individuals, it appears that no natural recruitment was evident in our samples. Genetic data were not able to clearly distinguish hatchery-bred stocked and wild-born fish. Understanding the effects of stocking on population composition is essential to evaluate the effectiveness of stocking to inform effective management and population recovery [1]. To date, the mark-recapture of chemically or physically tagged fish is the predominant method used to identify hatchery-bred stocked fish in Australia [2][3][4][5]. Otolith trace element composition has been widely applied in identifying the natal origin of fish [6][7][8][9][10][11]. This method has also been used to identify hatchery-bred stocked fish in wild salmonid populations [12][13][14].
We performed an otolith spot measurement to compare the chemical composition of the core (indicating natal origin) and edge (indicating current habitat). We observed that
Mg,
Sr,
Ba, and
Mn were the elements significantly contributing to the variation between core and edge composition. This is consistent with previous observations of otolith trace element composition in carp (
) from the Lachlan River [15].
In the upper Lachlan samples, no natural recruitment was evident using otolith microchemistry data. However, the clustering of the otolith edge, core, and hatchery reference groups was more complex than for the samples from the lower Lachlan. A different chemical signature was displayed by the core of otoliths compared to the edge as well as the YOY hatchery samples, indicating that the natal origin of these fish was neither the Lachlan River nor the Murray Darling Fisheries. Although our data could not conclusively identify the natal origin of these fish, plausible explanations for such results are (i) that they are fish stocked from hatcheries other than the Murray–Darling Fisheries such as DPI Fisheries (Figure 1) and (ii) that they are caused by interannual variability in the multi-element composition in the otolith, or a combination of both.
The fact that no association between high-flow events in the lower Lachlan and a priori recruitment events was evident in the present study suggests a flow-independent recruitment pattern in this species, which has also been reported in previous studies [16][17]. However, this could be an artifact of there being very few individuals from each spawning year. Larger sample sizes from each cohort will be required to provide conclusive evidence.
is a highly migratory, long-lived species and widely distributed in the MDB [18]. Within this region,
is considered as a single panmictic population with high gene flow across connected catchments, resulting in high genetic similarity among individuals from different drainages of the MDB [19][20]. Our approach to identify natal origin using SNP and SilicoDArT markers was unsuccessful. Such an outcome would be expected if (i) either one or both parents of the river-born fish were previously stocked hatchery-bred fish, which would indicate the natural recruitment of stocked golden perch in the Lachlan River; or (ii) the broodstock being used in the hatchery were previously stocked fish from a different cohort. The longevity of the species, the intensity of
stocking in Lachlan River, and the hatchery quality assurance program (the frequent replacement of old broodstock with wild-caught fish from the Lachlan River), coupled with abundant stocked fish in the river (the results from this study), suggests the second assumption is more likely to be correct. However, the natural recruitment in stocked fish is not infeasible considering a similar result in other species [21].
One possible limitation of this study that resulted in the inability of the genetic data to identify natal origin could be the low genomic representation of the hatchery samples. We sampled only a single cohort for the hatchery reference sample for the present study. Given the high genetic similarity of the species within the MDB as well as in Lachlan River, we recommend further analysis using multiple hatchery cohorts to demonstrate the effectiveness of genetic markers to identify hatchery-bred stocked and wild-born fish. The necessity of multiple-cohort sampling was also reflected in the otolith microchemistry analysis. We recommend further otolith study using multiple cohorts of hatchery reference as well as covering all the hatcheries contributing to the Lachlan River stocking. This will also reduce the probable error caused by physiological influences on otolith microchemistry, which was not considered in the present study [22][23].
There are increasing numbers of examples of studies that combine otolith microchemistry and genetic analyses [12][24][25][26][27]. As a combined approach, most studies have used otolith data to perform the fine-scale identification of spawning grounds and genetic data to investigate gene flow and connectivity among populations. Focusing on stock enhancement, we identified only one study where a similar research question to ours was addressed using a combination of genetic and otolith data. Perrier, Daverat, Evanno, Pécheyran, Bagliniere, and Roussel [12] used this multi-marker approach to infer information about the breeding activity of stocked
(Atlantic Salmon) in wild conditions. However, the utility of a multi-marker approach is dependent on the presence of fine-scale genetic differentiation between hatchery and wild fish. For genetically homogenized populations, otolith trace elements are the more useful approach to identify sub-populations from different localities (e.g., Svedäng, André, Jonsson, Elfman, and Limburg [27]).
In conclusion, the present study has found evidence of limited natural recruitment in the lower Lachlan River, while no natural recruitment was evident in the upper Lachlan. The study successfully demonstrates the utility of otolith microchemistry analysis to identify stocked and wild-born
- Crook, D.A.; O’Mahony, D.J.; Gillanders, B.M.; Munro, A.R.; Sanger, A.C.; Thurstan, S.; Baumgartner, L.J. Contribution of stocked fish to riverine populations of golden perch (Macquaria ambigua) in the Murray-Darling Basin, Australia. Mar. Freshw. Res. 2016, 67, 1401–1409. [Google Scholar] [CrossRef]
- Forbes, J.; Watts, R.J.; Robinson, W.A.; Baumgartner, L.J.; McGuffie, P.; Cameron, L.M.; Crook, D.A. Assessment of stocking effectiveness for Murray cod (Maccullochella peelii) and golden perch (Macquaria ambigua) in rivers and impoundments of south-eastern Australia. Mar. Freshw. Res. 2016, 67, 1410–1419. [Google Scholar] [CrossRef]
- Thiem, J.D.; Wooden, I.J.; Baumgartner, L.J.; Butler, G.L.; Forbes, J.P.; Conallin, J. Recovery from a fish kill in a semi-arid Australian river: Can stocking augment natural recruitment processes? Austral Ecol. 2017, 42, 218–226. [Google Scholar] [CrossRef]
- Amano, Y.; Kuwahara, M.; Takahashi, T.; Shirai, K.; Yamane, K.; Amakawa, H.; Otake, T. Otolith elemental and Sr isotopic composition as a natal tag for Biwa salmon Oncorhynchus masou subsp. in Lake Biwa, Japan. Aquat. Biol. 2013, 19, 85–95. [Google Scholar] [CrossRef]
- Artetxe-Arrate, I.; Fraile, I.; Crook, D.A.; Zudaire, I.; Arrizabalaga, H.; Greig, A.; Murua, H. Otolith microchemistry: A useful tool for investigating stock structure of yellowfin tuna (Thunnus albacares) in the Indian Ocean. Mar. Freshw. Res. 2019, 70, 1708–1721. [Google Scholar] [CrossRef]
- Ashford, J.; Arkhipkin, A.; Jones, C. Can the chemistry of otolith nuclei determine population structure of Patagonian toothfish Dissostichus eleginoides? J. Fish Biol. 2006, 69, 708–721. [Google Scholar] [CrossRef]
- Crook, D.; Macdonald, J.; O’Connor, J.; Barry, B. Use of otolith chemistry to examine patterns of diadromy in the threatened Australian grayling Prototroctes maraena. J. Fish Biol. 2006, 69, 1330–1344. [Google Scholar] [CrossRef]
- Ferguson, G.J.; Ward, T.M.; Gillanders, B.M. Otolith shape and elemental composition: Complementary tools for stock discrimination of mulloway (Argyrosomus japonicus) in southern Australia. Fish. Res. 2011, 110, 75–83. [Google Scholar] [CrossRef]
- Hearne, S.; Travers, M.; Evans, R.; Blyth, A.; Trinajstic, K.; McIlwain, J.; Newman, S. Population Connectivity of Two Reef Fish Species in Northwestern Australia Using Otolith Geochemistry: A Pilot Study; Report of Project 1.1.3—Project 1.1.3.4c Prepared for the Kimberley Marine Research Program; Western Australian Marine Science Institution: Perth, Western Australia, Australia, 2017. [Google Scholar]
- Coghlan, S.M., Jr.; Lyerly, M.S.; Bly, T.R.; Williams, J.S.; Bowman, D.; Hannigan, R. Otolith chemistry discriminates among hatchery-reared and tributary-spawned salmonines in a tailwater system. N. Am. J. Fish. Manag. 2007, 27, 531–541. [Google Scholar] [CrossRef]
- Olley, R.; Young, R.; Closs, G.P.; Kristensen, E.A.; Bickel, T.O.; Deans, N.; Davey, L.; Eggins, S. Recruitment sources of brown trout identified by otolith trace element signatures. N. Z. J. Mar. Freshw. Res. 2011, 45, 395–411. [Google Scholar] [CrossRef]
- Ebner, B.C.; Scholz, O.; Gawne, B. Golden perch Macquaria ambigua are flexible spawners in the Darling River, Australia. N. Z. J. Mar. Freshw. Res. 2009, 43, 571–578. [Google Scholar] [CrossRef]
- Attard, C.R.; Brauer, C.J.; Sandoval-Castillo, J.; Faulks, L.K.; Unmack, P.J.; Gilligan, D.M.; Beheregaray, L.B. Ecological disturbance influences adaptive divergence despite high gene flow in golden perch (Macquaria ambigua): Implications for management and resilience to climate change. Mol. Ecol. 2018, 27, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Beheregaray, L.B.; Pfeiffer, L.V.; Attard, C.R.; Sandoval-Castillo, J.; Domingos, F.M.; Faulks, L.K.; Gilligan, D.M.; Unmack, P.J. Genome-wide data delimits multiple climate-determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua). Mol. Phylogenet. Evol. 2017, 111, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Sturrock, A.M.; Hunter, E.; Milton, J.A.; Johnson, R.C.; Waring, C.P.; Trueman, C.N.; Eimf, E. Quantifying physiological influences on otolith microchemistry. Methods Ecol. Evol. 2015, 6, 806–816. [Google Scholar] [CrossRef]
- Sturrock, A.M.; Trueman, C.N.; Milton, J.A.; Waring, C.P.; Cooper, M.J.; Hunter, E. Physiological influences can outweigh environmental signals in otolith microchemistry research. Mar. Ecol. Prog. Ser. 2014, 500, 245–264. [Google Scholar] [CrossRef]
- Hughes, J.M.; Schmidt, D.J.; Macdonald, J.I.; Huey, J.A.; Crook, D.A. Low interbasin connectivity in a facultatively diadromous fish: Evidence from genetics and otolith chemistry. Mol. Ecol. 2014, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Reis-Santos, P.; Tanner, S.E.; Aboim, M.A.; Vasconcelos, R.P.; Laroche, J.; Charrier, G.; Pérez, M.; Presa, P.; Gillanders, B.M.; Cabral, H.N. Reconciling differences in natural tags to infer demographic and genetic connectivity in marine fish populations. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Schmidt, D.J.; Crook, D.A.; Macdonald, J.I.; Huey, J.A.; Zampatti, B.P.; Chilcott, S.; Raadik, T.A.; Hughes, J.M. Migration history and stock structure of two putatively diadromous teleost fishes, as determined by genetic and otolith chemistry analyses. Freshw. Sci. 2014, 33, 193–206. [Google Scholar] [CrossRef]
- Svedäng, H.; André, C.; Jonsson, P.; Elfman, M.; Limburg, K.E. Migratory behaviour and otolith chemistry suggest fine-scale sub-population structure within a genetically homogenous Atlantic Cod population. Environ. Biol. Fishes 2010, 89, 383–397. [Google Scholar] [CrossRef]
, particularly where a lack of genetic structure rules out other approaches. Along with the use of otoliths, we propose that parent–offspring assignment approaches with genetic data from the hatchery broodstock could be effective in terms of the genetic differentiation of stocked and wild-born fish.
References
- Taylor L. Hunt; Paul Jones; Informing the Great Fish Stocking Debate: An Australian Case Study. Reviews in Fisheries Science & Aquaculture 2018, 26, 275-308, 10.1080/23308249.2017.1407916.
- Taylor Hunt; Micheal S. Allen; John Douglas; Anne Gason; Evaluation of a Sport Fish Stocking Program in Lakes of the Southern Murray–Darling Basin, Australia. North American Journal of Fisheries Management 2010, 30, 805-811, 10.1577/m09-207.1.
- David A. Crook; Bronwyn M. Gillanders; Andrew Munro; Andrew C. Sanger; Stephen Thurstan; Lee J. Baumgartner; Damien J. O'mahony; Contribution of stocked fish to riverine populations of golden perch (Macquaria ambigua) in the Murray–Darling Basin, Australia. Marine and Freshwater Research 2016, 67, 1401-1409, 10.1071/mf15037.
- Jamin Forbes; Robyn J. Watts; Wayne A. Robinson; Lee J. Baumgartner; Prue McGuffie; Leo M. Cameron; David A. Crook; Assessment of stocking effectiveness for Murray cod (Maccullochella peelii) and golden perch (Macquaria ambigua) in rivers and impoundments of south-eastern Australia. Marine and Freshwater Research 2016, 67, 1410-1419, 10.1071/mf15230.
- Jason D. Thiem; Ian J. Wooden; Lee J. Baumgartner; Gavin L. Butler; Jamin Forbes; John Conallin; Recovery from a fish kill in a semi-arid Australian river: Can stocking augment natural recruitment processes?. Austral Ecology 2016, 42, 218-226, 10.1111/aec.12424.
- Y Amano; M Kuwahara; T Takahashi; Kotaro Shirai; K Yamane; H Amakawa; T Otake; Otolith elemental and Sr isotopic composition as a natal tag for Biwa salmon Oncorhynchus masou subsp. in Lake Biwa, Japan. Aquatic Biology 2013, 19, 85-95, 10.3354/ab00520.
- Artetxe-Arrate, I.; Fraile, I.; Crook, D.A.; Zudaire, I.; Arrizabalaga, H.; Greig, A.; Murua, H; Otolith microchemistry: A useful tool for investigating stock structure of yellowfin tuna (Thunnus albacares) in the Indian Ocean. Mar. Freshw. Res. 2019, 70, 1708–1721.
- Julian R. Ashford; A. I. Arkhipkin; C. M. Jones; Can the chemistry of otolith nuclei determine population structure of Patagonian toothfish Dissostichus eleginoides?. Journal of Fish Biology 2006, 69, 708-721, 10.1111/j.1095-8649.2006.01144.x.
- David A. Crook; J. I. Macdonald; B. Barry; J. P. O'connor; Use of otolith chemistry to examine patterns of diadromy in the threatened Australian grayling Prototroctes maraena. Journal of Fish Biology 2006, 69, 1330-1344, 10.1111/j.1095-8649.2006.01191.x.
- Greg Ferguson; Tim M. Ward; Bronwyn M. Gillanders; Otolith shape and elemental composition: Complementary tools for stock discrimination of mulloway (Argyrosomus japonicus) in southern Australia. Fisheries Research 2011, 110, 75-83, 10.1016/j.fishres.2011.03.014.
- Hearne, S.; Travers, M.; Evans, R.; Blyth, A.; Trinajstic, K.; McIlwain, J.; Newman, S. Population Connectivity of Two Reef Fish Species in Northwestern Australia Using Otolith Geochemistry: A Pilot Study; Report of Project 1.1.3—Project 1.1.3.4c Prepared for the Kimberley Marine Research Program; Western Australian Marine Science Institution: Perth, Western Australia, Australia, 2017.
- Charles Perrier; Françoise Daverat; Guillaume Evanno; Christophe Pécheyran; Jean-Luc Bagliniere; Jean-Marc Roussel; Coupling genetic and otolith trace element analyses to identify river-born fish with hatchery pedigrees in stocked Atlantic salmon (Salmo salar) populations. Canadian Journal of Fisheries and Aquatic Sciences 2011, 68, 977-987, 10.1139/f2011-040.
- Stephen M. Coghlan; Michael S. Lyerly; Thomas R. Bly; Jeffrey S. Williams; Darrell Bowman; Robyn Hannigan; Otolith Chemistry Discriminates among Hatchery-Reared and Tributary-Spawned Salmonines in a Tailwater System. North American Journal of Fisheries Management 2007, 27, 531-541, 10.1577/m06-118.1.
- R Olley; Roger Young; Gp Closs; Ea Kristensen; To Bickel; Na Deans; Ln Davey; Sm Eggins; Gerard Closs; Recruitment sources of brown trout identified by otolith trace element signatures. New Zealand Journal of Marine and Freshwater Research 2011, 45, 395-411, 10.1080/00288330.2011.592196.
- Macdonald, J.; McNeil, D.G.; Crook, D.A. Identification of Carp Recruitment Hotspots in the Lachlan River Using Otolith Chemistry; Report to the Invasive Animals CRC and Lachlan CMa; SaRDI Aquatic Sciences, Invasive Animals Cooperative Research Centre: Canberra, Australia, SARDI Publication No. F2009/000682-1. SARDI Research Report Series No. 434; 2010; Available online: https://www.researchgate.net/publication/247768728_Identification_of_Carp_Recruitment_Hotspots_in_the_Lachlan_River_Using_Otolith_Chemistry (accessed on 28 October 2019).
- Adam Kerezsy; S. R. Balcombe; Angela Arthington; Stuart E. Bunn; Continuous recruitment underpins fish persistence in the arid rivers of far-western Queensland, Australia. Marine and Freshwater Research 2011, 62, 1178-1190, 10.1071/mf11021.
- B. C. Ebner; O. Scholz; B. Gawne; Golden perch Macquaria ambigua are flexible spawners in the Darling River, Australia. New Zealand Journal of Marine and Freshwater Research 2009, 43, 571-578, 10.1080/00288330909510023.
- Lintermans, M. Fishes of the Murray-Darling Basin: An Introductory Guide; Murray-Darling Basin Commission: Canberra, Australia, 2007.
- Attard, C.R.; Brauer, C.J.; Sandoval-Castillo, J.; Faulks, L.K.; Unmack, P.J.; Gilligan, D.M.; Beheregaray, L.B; Ecological disturbance influences adaptive divergence despite high gene flow in golden perch (Macquaria ambigua): Implications for management and resilience to climate change. Mol. Ecol. 2018, 27, 196–215.
- Luciano B. Beheregaray; Lauren V. Pfeiffer; Catherine R. M. Attard; Jonathan Sandoval‐Castillo; Fabricius M. C. B. Domingos; Leanne K. Faulks; Dean Gilligan; Peter J Unmack; Genome-wide data delimits multiple climate-determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua). Molecular Phylogenetics and Evolution 2017, 111, 65-75, 10.1016/j.ympev.2017.03.021.
- Borwick, J.; District, A.; Philpot, A.; District, M.; Wilson, K. Review of Lake Simcoe’s Coldwater Fish Stocking Program; Ontario Ministry of Natural Resources: Midhurst (Aurora districts), ON, Canada, 2009.
- Anna M. Sturrock; Ewan Hunter; J. Andrew Milton; Rachel C. Johnson; Colin P. Waring; C. N. Trueman; Eimf; Eimf Eimf; Quantifying physiological influences on otolith microchemistry. Methods in Ecology and Evolution 2015, 6, 806-816, 10.1111/2041-210x.12381.
- Am Sturrock; Cn Trueman; Ja Milton; Cp Waring; Mj Cooper; Ewan Hunter; Physiological influences can outweigh environmental signals in otolith microchemistry research. Marine Ecology Progress Series 2014, 500, 245-264, 10.3354/meps10699.
- J. M. Hughes; Dan Schmidt; Jed I. Macdonald; Joel A. Huey; David A. Crook; Low interbasin connectivity in a facultatively diadromous fish: evidence from genetics and otolith chemistry. Molecular Ecology 2014, 23, 1000-1013, 10.1111/mec.12661.
- Patrick Reis-Santos; Susanne E Tanner; Maria Ana Aboim; Rita P. Vasconcelos; Jean Laroche; Grégory Charrier; Montse Pérez; Pablo Presa; Bronwyn M. Gillanders; Henrique N. Cabral; et al. Reconciling differences in natural tags to infer demographic and genetic connectivity in marine fish populations. Scientific Reports 2018, 8, 1–12, 10.1038/s41598-018-28701-6.
- Dan Schmidt; David A. Crook; Jed I. Macdonald; Joel A. Huey; Brenton P. Zampatti; Stuart Chilcott; Tarmo A. Raadik; J. M. Hughes; Migration history and stock structure of two putatively diadromous teleost fishes, as determined by genetic and otolith chemistry analyses. Freshwater Science 2014, 33, 193-206, 10.1086/674796.
- Henrik Svedäng; Carl André; Patrik Jonsson; Mikael Elfman; Karin E. Limburg; Migratory behaviour and otolith chemistry suggest fine-scale sub-population structure within a genetically homogenous Atlantic Cod population. Journal of Applied Phycology 2010, 89, 383-397, 10.1007/s10641-010-9669-y.
