Aurora kinase B (AURKB) is a mitotic serine/threonine protein kinase that belongs to the aurora kinase family along with aurora kinase A (AURKA) and aurora kinase C (AURKC). AURKB is a member of the chromosomal passenger protein complex and plays a role in cell cycle progression.
- aurora kinase B (AURKB)
- cancer
- AURKB regulation
- tumors
1. Introduction
Aurora kinases (AURKs) are protein serine/threonine kinases consisting of three members in the gene family—aurora kinase A (AURKA), aurora kinase B (AURKB) and aurora kinase C (AURKC) [1]. AURKs are critical regulators of the cell cycle, with AURKA and AURKB playing a key function in mitosis [1], whereas AURKC has a significant role in gametogenesis [2,3][2][3]. AURKs have three different domains, of which the kinase domain shares a high degree of homology among all the members [4]. The functions of AURKs are well-defined by their localization and spatio-temporal expression [5] and also the sequence differences in their N-terminal region [4]. Overexpression of AURKs in tumors has been shown to trigger aneuploidy and genomic instability [6] that leads to tumor development, invasion and metastasis. AURKA and its functions have been widely investigated and several reviews were authored to highlight its importance in cancer [7,8][7][8]. The role of AURKC in cancer is not completely understood and needs further investigations. This review focuses on AURKB, a potential drug target from the AURK gene family.
Aurora kinase B is encoded by the AURKB gene located on chromosome 17 and is also known by other names including AIK2, AIM1, ARK2, AIRK2, IPL1, STK1, STK5 and STK12. AURKB along with other AURKs plays a vital role in the regulation of cell cycle. Phosphorylation of histone H3 by aurora kinases is essential for chromosome segregation during cell division. Both AURKA and AURKB have been shown to phosphorylate histone H3 [9]. The role of elevated AURKB in increasing the phosphorylation of histone H3 on Ser10 and aneuploidy has been elucidated in studies involving Chinese hamster embryo cells exogenously overexpressing AURKB. The function of AURKB in transformation was further demonstrated in vivo by injecting these AURKB overexpressing cells into BALB/c nu/nu mice [10]. Induced AURKB expression is also linked to tumorigenesis mediated by H-Ras [11].
2. Structure and Function of AURKB
AURKB, similar to other members of the aurora kinase gene family, is composed of three domains: (i) N-terminal domain, (ii) kinase domain, and (iii) C–terminal domain [12]. The kinase domain or catalytic domain is highly conserved between all the three members of the aurora family, whereas the N-terminal domain shows a varied degree of sequence dissimilarity that provides selectivity for protein-protein interactions. The kinase domain in AURKB is composed of a β-stranded lobe on the N-terminal side and an α-helical lobe on the C-terminal side. These two lobes are connected together by a hinge region, which permits active kinase conformation [13]. The C-terminal lobe of the kinase domain contains the catalytic T-loop whose auto-phosphorylation at Thr232 results in AURKB activation. AURKB contains three types of degrons; KEN motif, D-box and DAD/A box that are thought to mediate its degradation. The catalytic domain contains D-boxes (reviewed in [14]). The non-catalytic N-terminal domain has a KEN motif and a DAD/A box, whereas the C-terminal domain has a D-box. However, Nguyen et al., has showed that only N-terminal KEN and DAD/A boxes are responsible for AURKB degradation [15] (Figure 1a).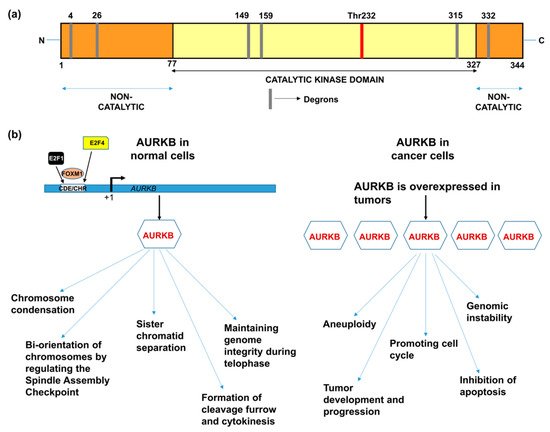
3. Deregulation of AURKB in Cancer
Drawing parallels with the functions of AURKB in mitosis, it is anticipated that alterations of AURKB either as amplification or overexpression could provide a proliferative advantage to cancer cells (Figure 1b). In fact, its role in tumor cell transformation was clarified by overexpressing AURKB in murine epithelial cells. When these AURKB overexpressing cells were injected into nude mice, formation of mammary epithelial tumors was observed along with amplifications and deletions of DNA isolated from these mouse mammary tumors [26]. Over the years, aberrant AURKB expression has been reported in a variety of malignancies including human seminoma [27], and thyroid carcinoma [28]. Using mRNA expression analysis Smith et al., found that AURKB expression is markedly elevated in non-small cell lung carcinoma (NSCLC) in comparison to matched untransformed lung tissues. This study also showed that AURKB overexpression led to poor progression free survival, however overall survival was not affected [29]. In contrast to this finding, a study by Vischioni et al., has found that AURKB expression is significantly associated with older age at diagnosis and reduced overall survival in adenocarcinoma subtype of NSCLC [30]. Likewise, AURKB was found to be induced in the squamous carcinoma subtype of NSCLC and the expression level of AURKB also served as a marker for resistance to paclitaxel, a drug commonly used in the treatment of NSCLC [31]. Similar to NSCLC, metastatic colorectal cancer patients lived significantly shorter when they had high levels of AURKB expression in their tumor tissues [32] and the publicly available databases further showed that increased AURKB expression also correlated significantly with reduced survival in breast cancer patients [33]. AURKB expression has been identified as a prognostic biomarker in glioblastoma [34], gastric cancer [35] and oral cancer [36,37,38][36][37][38]. AURKB overexpression was found to be increased in prostate cancer tissues compared to healthy controls [39]. Hepatocellular carcinoma tissues also showed significantly elevated mRNA expression of AURKB compared to paired healthy liver tissues and was found to be an independent prognostic marker for tumor invasiveness and prognosis [40]. Pediatric acute lymphoblastic leukemia and acute myeloid leukemia (AML) exhibited high levels of both AURKA and AURKB compared to control bone marrow mononuclear cells; however, the inhibition of AURKB alone resulted in apoptosis suggesting that AURKB is a putative drug target but not AURKA in these hematologic malignancies [41]. Recently, we have shown that AURKB is overexpressed in retinoblastoma compared to adjacent healthy retina and its expression significantly correlated with histological risk factors such as optic nerve and anterior chamber invasion [42]. Additionally, we have retrieved expression data of AURKB from Gene Expression Profiling Interactive Analysis (GEPIA) database [43] for a variety of cancers and compared it to their respective normal tissue counterparts (Figure 2). The above studies demonstrating the overexpression of AURKB in tumors compared to their non-cancerous counterpart tissues suggest the possible therapeutic targeting of AURKB in tumors.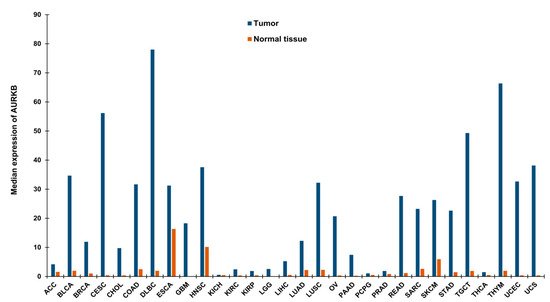
4. Targeting AURKB in cancer
Deregulated expression of AURKs is increasingly viewed as a potential drug target. AURKs are successfully inhibited in several preclinical cell line and animal models. A variety of small molecule inhibitors to target AURKs have been developed and are in different phases of clinical trials. The development of AURKA inhibitors and their progress has been described recently [7]. The possibility of targeting AURKB has gained momentum during last few years.
Taking into account that the members of the aurora kinase family have very high homology in the kinase domain, most small molecule inhibitors developed against aurora kinases have overlapping inhibitory activity. Despite this, pioneering collaborations between industry and academia led to the development of Hesperadin and ZM447439, the former being predominantly selective for AURKB. ZM447439 was further modified to develop AZD1152. Most of the inhibitors were synthesized with an aim to improve therapy, but over the years, they have been extensively used to understand the varied functions and complex regulations mediated by AURKB.
5. Conclusion
Aurora kinase B is gaining prominence as a therapeutic target in a variety of cancers. Currently, there are many small molecule inhibitors (Barasertib, Danusertib, etc.) that have been tested in clinical trials. Further, nanoparticle formulations of AURKB inhibitors have been designed to improve efficacy. In summary, AURKB inhibition seems to be a promising strategy for combating tumors.
References
- Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell. Biol. 2001, 2, 21–32.
- Dieterich, K.; Soto Rifo, R.; Karen Faure, A.; Hennebicq, S.; Amar, B.B.; Zahi, M.; Perrin, J.; Martinez, D.; Sèle, B.; Jouk, P.S.; et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat. Genet. 2007, 39, 661–665.
- Sharif, B.; Na, J.; Lykke-Hartmann, K.; McLaughlin, S.H.; Laue, E.; Glover, D.M.; Zernicka-Goetz, M. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J. Cell Sci. 2010, 123, 4292–4300.
- Carmena, M.; Earnshaw, W.C. The cellular geography of Aurora kinases. Nat. Rev. Mol. Cell Biol. 2003, 4, 842–854.
- Li, S.; Deng, Z.; Fu, J.; Xu, C.; Xin, G.; Wu, Z.; Luo, J.; Wang, G.; Zhang, S.; Zhang, B.; et al. Spatial compartmentalization specializes the function of Aurora A and Aurora B. J. Biol. Chem. 2015, 290, 17546–17558.
- Marumoto, T.; Zhang, D.; Saya, H. Aurora-A—A guardian of poles. Nat. Rev. Cancer 2005, 5, 42–50.
- Galetta, D.; Cortes-Dericks, L. Promising therapy in lung cancer: Spotlight on aurora kinases. Cancers 2020, 12, 3371.
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora kinases: Novel therapy targets in cancers. Oncotarget 2017, 8, 23937–23954.
- Crosio, C.; Fimia, G.M.; Loury, R.; Kimura, M.; Okano, Y.; Zhou, H.; Sen, S.; Allis, C.D.; Sassone-corsi, P. Mitotic Phosphorylation of Histone H3: Spatio-Temporal Regulation by Mammalian Aurora Kinases. Mol. Cell. Biol. 2002, 22, 874–885.
- Ota, T.; Suto, S.; Katayama, H.; Han, Z.B.; Suzuki, F.; Maeda, M.; Tanino, M.; Terada, Y.; Tatsuka, M. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002, 62, 5168–5177.
- Kanda, A.; Kawai, H.; Suto, S.; Kitajima, S.; Sato, S.; Takata, T.; Tatsuka, M. Aurora-B/AIM-1 kinase activity is involved in Ras-mediated cell transformation. Oncogene 2005, 24, 7266–7272.
- Bolanos-Garcia, V.M. Aurora kinases. Int. J. Biochem. Cell Biol. 2005, 37, 1572–1577.
- Cheetham, G.M.T.; Knegtel, R.M.A.; Coll, J.T.; Renwick, S.B.; Swenson, L.; Weber, P.; Lippke, J.A.; Austen, D.A. Crystal structure of Aurora-2, an oncogenic serine/threonine kinase. J. Biol. Chem. 2002, 277, 42419–42422.
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The functional diversity of Aurora kinases: A comprehensive review. Cell Div. 2018, 1–17.
- Nguyen, H.G.; Chinnappan, D.; Urano, T.; Ravid, K. Mechanism of Aurora-B Degradation and Its Dependency on Intact KEN and A-Boxes: Identification of an Aneuploidy-Promoting Property. Mol. Cell. Biol. 2005, 25, 4977–4992.
- Portella, G.; Passaro, C.; Chieffi, P. Aurora B: A New Prognostic Marker and Therapeutic Target in Cancer. Curr. Med. Chem. 2011, 18, 482–496.
- Vader, G.; Medema, R.H.; Lens, S.M.A. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J. Cell Biol. 2006, 173, 833–837.
- Gassmann, R.; Carvalho, A.; Henzing, A.J.; Ruchaud, S.; Hudson, D.F.; Honda, R.; Nigg, E.A.; Gerloff, D.L.; Earnshaw, W.C. Borealin: A novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004, 166, 179–191.
- Zeitlin, S.G.; Shelby, R.D.; Sullivan, K.F. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001, 155, 1147–1157.
- Broad, A.J.; DeLuca, K.F.; DeLuca, J.G. Aurora B kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J. Cell Biol. 2020, 219.
- Yi, Q.; Chen, Q.; Yan, H.; Zhang, M.; Liang, C.; Xiang, X.; Pan, X.; Wang, F. Aurora B kinase activity– dependent and –independent functions of the chromosomal passenger complex in regulating sister chromatid cohesion. J. Biol. Chem. 2019, 294, 2021–2035.
- Uehara, R.; Tsukada, Y.; Kamasaki, T.; Poser, I.; Yoda, K.; Gerlich, D.W.; Goshima, G. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 2013, 202, 623–636.
- Schumacher, J.M.; Golden, A.; Donovan, P.J. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998, 143, 1635–1646.
- Giet, R.; Glover, D.M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001, 152, 669–682.
- Adams, R.R.; Maiato, H.; Earnshaw, W.C.; Carmena, M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001, 153, 865–880.
- Nguyen, H.G.; Makitalo, M.; Yang, D.; Chinnappan, D.; St.Hilaire, C.; Ravid, K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009.
- Chieffi, P.; Troncone, G.; Caleo, A.; Libertini, S.; Linardopoulos, S.; Tramontano, D.; Portella, G. Aurora B expression in normal testis and seminomas. J. Endocrinol. 2004, 181, 263–270.
- Sorrentino, R.; Libertini, S.; Pallante, P.L.; Troncone, G.; Palombini, L.; Bavetsias, V.; Spalletti-Cernia, D.; Laccetti, P.; Linardopoulos, S.; Chieffi, P.; et al. Aurora B overexpression associates with the thyroid carcinoma undifferentiated phenotype and is required for thyroid carcinoma cell proliferation. J. Clin. Endocrinol. Metab. 2005, 90, 928–935.
- Smith, S.L.; Bowers, N.L.; Betticher, D.C.; Gautschi, O.; Ratschiller, D.; Hoban, P.R.; Booton, R.; Santibáñez-Koref, M.F.; Heighway, J. Overexpression of aurora B kinase (AURKB) in primary non-small cell lung carcinoma is frequent, generally driven from one allele, and correlates with the level of genetic instability. Br. J. Cancer 2005, 93, 719–729.
- Vischioni, B.; Oudejans, J.J.; Vos, W.; Rodriguez, J.A.; Giaccone, G. Frequent overexpression of aurora B kinase, a novel drug target, in non-small cell lung carcinoma patients. Mol. Cancer Ther. 2006, 5, 2905–2913.
- Al-Khafaji, A.S.K.; Davies, M.P.A.; Risk, J.M.; Marcus, M.W.; Koffa, M.; Gosney, J.R.; Shaw, R.J.; Field, J.K.; Liloglou, T. Aurora B expression modulates paclitaxel response in non-small cell lung cancer. Br. J. Cancer 2017, 116, 592–599.
- Pohl, A.; Azuma, M.; Zhang, W.; Yang, D.; Ning, Y.; Winder, T.; Danenberg, K.; Lenz, H.J. Pharmacogenetic profiling of Aurora kinase B is associated with overall survival in metastatic colorectal cancer. Pharm. J. 2011, 11, 93–99.
- Huang, D.; Huang, Y.; Huang, Z.; Weng, J.; Zhang, S.; Gu, W. Relation of AURKB over-expression to low survival rate in BCRA and reversine-modulated aurora B kinase in breast cancer cell lines. Cancer Cell Int. 2019, 19, 1–13.
- Zeng, W.F.; Navaratne, K.; Prayson, R.A.; Weil, R.J. Aurora B expression correlates with aggressive behaviour in glioblastoma multiforme. J. Clin. Pathol. 2007, 60, 218–221.
- Nie, M.; Wang, Y.; Yu, Z.; Li, X.; Deng, Y.; Wang, Y.; Yang, D.; Li, Q.; Zeng, X.; Ju, J.; et al. AURKB promotes gastric cancer progression via activation of CCND1 expression. Aging 2020, 12, 1304–1321.
- Qi, G.; Ogawa, I.; Kudo, Y.; Miyauchi, M.; Siriwardena, B.S.M.S.; Shimamoto, F.; Tatsuka, M.; Takata, T. Aurora-B expression and its correlation with cell proliferation and metastasis in oral cancer. Virchows Arch. 2007, 450, 297–302.
- Pannone, G.; Hindi, S.A.H.; Santoro, A.; Sanguedolce, F.; Rubini, C.; Cincione, R.I.; De Maria, S.; Tortorella, S.; Rocchetti, R.; Cagiano, S.; et al. Aurora B expression as a prognostic indicator and possibile therapeutic target in oral squamous cell carcinoma. Int. J. Immunopathol. Pharmacol. 2011, 24, 79–88.
- Li, M.; Liu, H.; Zhao, Q.; Han, S.; Zhou, L.; Liu, W.; Li, W.; Gao, F. Targeting Aurora B kinase with Tanshinone IIA suppresses tumor growth and overcomes radioresistance. Cell Death Dis. 2021, 12.
- Chieffi, P.; Cozzolino, L.; Kisslinger, A.; Libertini, S.; Staibano, S.; Mansueto, G.; De Rosa, G.; Villacci, A.; Vitale, M.; Linardopoulos, S.; et al. Aurora B expression directly correlates with prostate cancer malignancy and influence prostate cell proliferation. Prostate 2006, 66, 326–333.
- Lin, Z.Z.; Jeng, Y.M.; Hu, F.C.; Pan, H.W.; Tsao, H.W.; Lai, P.L.; Lee, P.H.; Cheng, A.L.; Hsu, H.C. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer 2010, 10.
- Hartsink-Segers, S.A.; Zwaan, C.M.; Exalto, C.; Luijendijk, M.W.J.; Calvert, V.S.; Petricoin, E.F.; Evans, W.E.; Reinhardt, D.; De Haas, V.; Hedtjärn, M.; et al. Aurora kinases in childhood acute leukemia: The promise of aurora B as therapeutic target. Leukemia 2013, 27, 560–568.
- Borah, N.A.; Sradhanjali, S.; Barik, M.R.; Jha, A.; Tripathy, D.; Kaliki, S.; Rath, S.; Raghav, S.K.; Patnaik, S.; Mittal, R.; et al. Aurora Kinase B Expression, Its Regulation and Therapeutic Targeting in Human Retinoblastoma. Investig. Ophthalmol. Vis. Sci. 2021, 62, 16.
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102.
