In the wake of the COVID-19 pandemic, it is crucial to assess the application of a multitude of effective diagnostic specimens for conducting mass testing, for accurate diagnosis and to formulate strategies for its prevention and control. As one of the most versatile and amenable specimen options, saliva offers great advantages for widespread screening strategies due to its non-invasive properties, cost-effectiveness, excellent stability and minimal risk of cross-infection.
- saliva
- COVID-19
- SARS-CoV-2
- infection
- diagnosis
- polymerase chain reaction
1. Introduction
SARS-CoV-2 can be transmitted via direct or indirect contact. One of the primary sources of transmission of coronavirus is through salivary aerosols emitted from coughing, breathing, and even during speaking [2].
SARS-CoV-2 can be transmitted via direct or indirect contact. One of the primary sources of transmission of coronavirus is through salivary aerosols emitted from coughing, breathing, and even during speaking [1].
Figure 1, below, illustrates three potential trajectories for the presence of the virus in saliva as explained by Sabino-Silva et al. [3]. Contemporary studies have gathered evidence demonstrating molecular strategies adopted by the SARS-CoV-2 virus, enabling it to enter the host cell, causing a high rate of infectivity. Possible activation of the SARS-CoV-2 virus by gene expression of furin in salivary glands is also a noteworthy finding explained by Shang et al. [4]. Furin is typically expressed by salivary glands and its components are responsible for the regulation of different specific proteins while the gene itself is believed to cleave different viral toxins including coronaviruses. As a result, the severity of COVID-19 disease is increased if salivary infection is withdrawn from the salivary glands, while the presence of furin in saliva leads to a rapid progression of the disease through salivary droplets [5].
, below, illustrates three potential trajectories for the presence of the virus in saliva as explained by Sabino-Silva et al. [2]. Contemporary studies have gathered evidence demonstrating molecular strategies adopted by the SARS-CoV-2 virus, enabling it to enter the host cell, causing a high rate of infectivity. Possible activation of the SARS-CoV-2 virus by gene expression of furin in salivary glands is also a noteworthy finding explained by Shang et al. [3]. Furin is typically expressed by salivary glands and its components are responsible for the regulation of different specific proteins while the gene itself is believed to cleave different viral toxins including coronaviruses. As a result, the severity of COVID-19 disease is increased if salivary infection is withdrawn from the salivary glands, while the presence of furin in saliva leads to a rapid progression of the disease through salivary droplets [4].
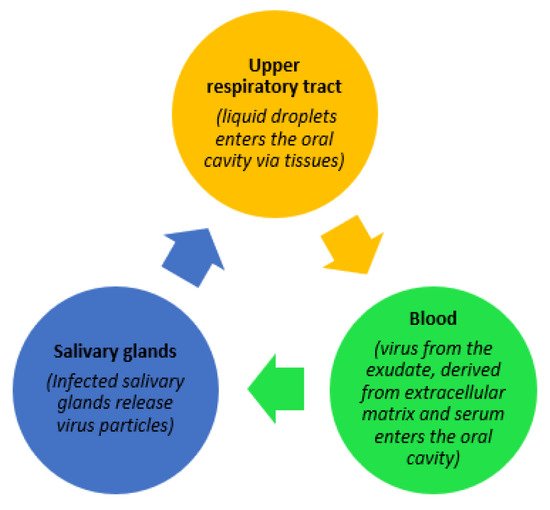
Figure 1.
Possible trajectories for the presence of SARS-CoV-2 in saliva.
2. Salivary Diagnostics
Table 2 describes assays that have received FDA EUA approval containing more recent data. Different approaches and collection techniques were used in the trials included here, such as collection of saliva by cough, passive collection from posterior oro-pharynx, simple swab or a whole saliva collection technique. As of 22 July 2020, RT-PCR using nasopharyngeal respiratory specimens is very much the gold standard for the qualitative detection of the SARS-COV-2 virus.
1 describes assays that have received FDA EUA approval containing more recent data. Different approaches and collection techniques were used in the trials included here, such as collection of saliva by cough, passive collection from posterior oro-pharynx, simple swab or a whole saliva collection technique. As of 22 July 2020, RT-PCR using nasopharyngeal respiratory specimens is very much the gold standard for the qualitative detection of the SARS-COV-2 virus.
Comparison of studies using saliva-based testing versus conventional swab-based testing for the detection of SARS-COV-2.
| Study | Ref | Saliva Collection Method | Swabs and Lavage for Comparison | Diagnostic Test | N | TP | FP | FN | TN | Sensitivity | Specificity | PPV | NPV |
|---|
| Azzi, L et al., 2020 (Italy) | [12] | [5] | Drooling | NPS | RT-PCR | 25 | 25 | 0 | 0 | 0 | 1 | uc | 1 | uc |
| Azzi, L et al., 2020 (Italy) | [13] | [6] | Drooling | BAL | RT-PCR | 2 | 0 | 2 | 0 | 0 | uc | 0 | 0 | uc |
| Chen, Lili et al., 2020 (China) | [14] | [7] | Cotton Swabs—Saliva from orifices | OPS | RT-qPCR | 31 | 4 | 0 | 9 | 18 | 0.31 | 1 | 1 | 0.66 |
| Han, Mi Seon et al., 2020 (Korea) | [15] | [8] | Saliva | NPS, OPS | qPCR | 2 | 1 | 0 | 1 | 0 | 0.50 | uc | 1 | 0 |
| Wang, To et al., 2020 (Hong Kong, China) | [16] | [9] | Sputum/Coughed-out Saliva (self-collected) | NPS | RT-qPCR | 12 | 11 | 0 | 1 | 0 | 0.92 | uc | 1 | 0 |
| Wang, To et al., 2020 (Hong Kong, China) | [17] | [10] | Coughed-up Saliva—Posterior OroPharynx | NPS, Sputum | RT-qPCR | 23 | 20 | 0 | 3 | 0 | 0.87 | uc | 1 | 0 |
| Wyllie Anne et al., 2020 (USA) | [18] | [11] | Saliva (spitting) | NPS | rRT-PCR | 46 | 38 | 1 | 7 | 0 | 0.84 | 0 | 0.97 | 0 |
| Zheng Shufa et al., 2020 (China) | [19] | [12] | Sputum (hospitalized patients) | Stool, Serum, Urine | RT-qPCR | 96 | 96 | 0 | 0 | 0 | 1 | uc | 1 | uc |
| Zhang Wei et al., 2020 (China) | [20] | [13] | Oral Swabs (hospitalized patients—baseline) | Blood, Anal | RT-qPCR | 16 | 8 | 0 | 8 | 0 | 0.50 | uc | 1 | 0 |
| Pasomsub, E et al., 2020 (Thailand) | [21] | [14] | Saliva | NPS, TS | RT-PCR | 200 | 16 | 2 | 3 | 179 | 0.84 | 0.98 | 0.88 | 0.98 |
| Somrak et al., 2021 | [22] | [15] | Self-collected | NPS | RT-PCR | 32 | 12 | 0 | 20 | 0 | 0.37 | 1 | 1 | 0.91 |
| Basso et al., 2021 | [23] | [16] | Self-collected | NPS | RT-PCR | 84 | 67 | 0 | 17 | 0 | 0.78 | uc | 1 | 0 |
A study in Hong Kong is the earliest available reported study during the course of pandemic, which investigated the presence of SARS-CoV-2 in saliva in 11 COVID-19-positive patients. The patients were tested at various phases including during their recovery phase and, at that time, a decline in salivary SARS-CoV-2 RNA was observed [24][17]. Early evidence from Wuhan (China), revealed that in a cohort of 16 COVID-19 patients, the SARS-CoV-2 viral titers were discovered using oral swabs, anal swabs and in plasma; but investigators also found that the detection overlap between the three samples was not consistent. Further investigations revealed positive oral swab results in eight patients following medical treatment [20][13]. Important studies evaluated the range of viral loads present in saliva and researchers found values ranging from 9.9 × 102 to 1.2 × 108 copies/mL [17,25,26,27,28,29,30,31,32][10][18][19][20][21][22][23][24][25]. Other investigators evaluated and compared the difference in efficiency of saliva using oro-nasopharyngeal swabs for the detection of viral load. Other studies validated the sensitivity of saliva samples versus nasopharyngeal swabs using RT-qPCR analysis and these are reported in multiple studies [12,21,30,32,33,34,35,36,37][5][14][23][25][26][27][28][29][30]. In a case study on a COVID-19-infected neonate, Korean investigators identified that serial sampling of saliva demonstrated a reduction in viral load over a 27-day follow-up [15][8]. Historically, perhaps, the first study that investigated the clinical progression of the disease course with temporal viral load also confirmed that posterior oropharyngeal salivary viral load was highest in the first week after symptom onset, and subsequently declined with time [17][10]. In a separate study, investigators from the USA compared the sensitivity and specificity of nasopharyngeal swabs versus saliva samples for the detection of SARS-CoV-2 via RT-PCR and demonstrated that saliva had a higher detection sensitivity, maintained consistency throughout the course of infection, and demonstrated less variability during the self-sampling collection process [18][11]. Furthermore, in a case report of two patients from an Italian study, investigators demonstrated the positive detection of SARS-COV-2 virus in saliva specimens, while respiratory swab specimens indicated a negative result in both cases [13][6]. In a study by Wong et al., the cost of these two specimens was compared and it was estimated that saliva specimens (USD 8.24 per 100) were much more economical when compared to the use of NPS (USD 104.87 per 100) [38][31].
3. Sensitivity and Specificity of Salivary Diagnostics for SARS-CoV-2 Testing
Limited studies have demonstrated the comparability or superiority of saliva sampling, relative to conventional swab-based sampling; however, the results for saliva are compelling. As mentioned earlier, we were able to compile the results from current and previously reported studies and have been able to calculate a pooled sensitivity and specificity for saliva specimen collection in the detection of SARS-CoV-2. The table below (see
Table 2) demonstrates a pooled sensitivity of approximately 87% and a specificity of 98%. From the combined results, the probability of a positive test result being a true positive (PPV/True-positive) is 98%, and the probability of a negative test result being a true negative result (NPV/False-positive) is 86%. These findings indicate that salivary-based detection of SARS-CoV-2 using the RT-PCR method has a high diagnostic accuracy for positive cases but may lack accuracy for the detection of false-positive cases. However, these performance data mirror that of data obtained using nasopharyngeal swabs and represents a solid alternative for diagnostic purposes.
1) demonstrates a pooled sensitivity of approximately 87% and a specificity of 98%. From the combined results, the probability of a positive test result being a true positive (PPV/True-positive) is 98%, and the probability of a negative test result being a true negative result (NPV/False-positive) is 86%. These findings indicate that salivary-based detection of SARS-CoV-2 using the RT-PCR method has a high diagnostic accuracy for positive cases but may lack accuracy for the detection of false-positive cases. However, these performance data mirror that of data obtained using nasopharyngeal swabs and represents a solid alternative for diagnostic purposes.
3.1. Strengths and Limitations of Salivary Diagnostics
The inclusion of saliva samples for the detection of SARS-CoV-2 coronavirus is a major step forward in the fight to identify patients suffering from the disease. Due to its easily accessible nature, it can be readily obtained from patients in a non-invasive fashion, thereby reducing the risk of nosocomial infections among healthcare workers [8,54]. Equally importantly, saliva samples possess high sensitivity and specificity when compared to nasopharyngeal swabs for the detection of SARS-CoV-2 coronavirus (refer to
The inclusion of saliva samples for the detection of SARS-CoV-2 coronavirus is a major step forward in the fight to identify patients suffering from the disease. Due to its easily accessible nature, it can be readily obtained from patients in a non-invasive fashion, thereby reducing the risk of nosocomial infections among healthcare workers [32][33]. Equally importantly, saliva samples possess high sensitivity and specificity when compared to nasopharyngeal swabs for the detection of SARS-CoV-2 coronavirus (refer to
Table 2). Intriguingly, there are also a few studies that report that SARS-CoV-2 is detectable in saliva samples but not in nasopharyngeal swabs [13,18]. It is documented that saliva collection is beneficial in cases where screening of infected individuals is required on a larger scale such as in the community, in a drive-through setting, in a hospital setup or in locations with access to limited medical resources. In addition, it has also been previously documented that certain viral strains may survive in saliva for 29 days post-infection, enhancing the possibility of disease detection even at later stages of the disease [3].
1). Intriguingly, there are also a few studies that report that SARS-CoV-2 is detectable in saliva samples but not in nasopharyngeal swabs [6][11]. It is documented that saliva collection is beneficial in cases where screening of infected individuals is required on a larger scale such as in the community, in a drive-through setting, in a hospital setup or in locations with access to limited medical resources. In addition, it has also been previously documented that certain viral strains may survive in saliva for 29 days post-infection, enhancing the possibility of disease detection even at later stages of the disease [2].
3.2. Emerging Technologies in Salivary Diagnostics for COVID-19
Besides the CDC-approved RT-PCR test for the detection of SARS-COV-2, many other inexpensive, fast detection methods for mass screening purposes have been recently approved by the FDA under the EUA (Emergency Use Authorization) mechanism [10]. As illustrated in
Besides the CDC-approved RT-PCR test for the detection of SARS-COV-2, many other inexpensive, fast detection methods for mass screening purposes have been recently approved by the FDA under the EUA (Emergency Use Authorization) mechanism [34]. As illustrated in
, the diagnostic specificity and sensitivity of oral fluids (saliva and sputum) are very high in controlled studies with smaller sample sizes.
4. Direction for Future Studies
Large-scale prospective studies are needed to establish the temporal trends in salivary viral titers and tie them in with the course of infection or as markers of disease severity. Although preliminary evidence indicates that salivary detection of SARS-CoV-2 is feasible in mild or asymptomatic cases [16,17], once again this finding needs to be validated in a larger cohort. Large-scale, epidemiological studies are needed to compare the sensitivity and specificity of swab-based methods versus methods where saliva specimens are used. Here we note that there are differences in the literature where various types of oral samples collected have contributed to variability in the detection of SARS-CoV-2 (refer to
Large-scale prospective studies are needed to establish the temporal trends in salivary viral titers and tie them in with the course of infection or as markers of disease severity. Although preliminary evidence indicates that salivary detection of SARS-CoV-2 is feasible in mild or asymptomatic cases [9][10], once again this finding needs to be validated in a larger cohort. Large-scale, epidemiological studies are needed to compare the sensitivity and specificity of swab-based methods versus methods where saliva specimens are used. Here we note that there are differences in the literature where various types of oral samples collected have contributed to variability in the detection of SARS-CoV-2 (refer to
Table 2). This in turn leads us to recommend that further studies should be performed to validate different oral fluid collection protocols and to compare viral detection rates [20,55]. Moreover, it is important to extend the application of salivary diagnostics to neglected or vulnerable populations such as pediatric populations, geriatrics and pregnant females. Generally speaking, the literature is unclear on whether SARS-CoV-2 detection is dependent on ACE-2 receptor expression at oral sites, versus nasopharyngeal sites, so we believe this relationship should be investigated in future studies [56]. To avoid the frequency of false-negative results in suspected positive cases, sampling a mix of multiple specimens (including oropharyngeal, nasopharyngeal, oral, sputum and saliva specimens) is recommended [57]. For proposed future research, we recommend the application of saliva for a number of studies aimed at disease detection and looking at progression. Saliva is an excellent matrix for the evaluation of salivary antibodies, so studies on a large cohort of individuals will provide advantages for monitoring disease progression in the COVID-19 area.
1). This in turn leads us to recommend that further studies should be performed to validate different oral fluid collection protocols and to compare viral detection rates [13][35]. Moreover, it is important to extend the application of salivary diagnostics to neglected or vulnerable populations such as pediatric populations, geriatrics and pregnant females. Generally speaking, the literature is unclear on whether SARS-CoV-2 detection is dependent on ACE-2 receptor expression at oral sites, versus nasopharyngeal sites, so we believe this relationship should be investigated in future studies [36]. To avoid the frequency of false-negative results in suspected positive cases, sampling a mix of multiple specimens (including oropharyngeal, nasopharyngeal, oral, sputum and saliva specimens) is recommended [37]. For proposed future research, we recommend the application of saliva for a number of studies aimed at disease detection and looking at progression. Saliva is an excellent matrix for the evaluation of salivary antibodies, so studies on a large cohort of individuals will provide advantages for monitoring disease progression in the COVID-19 area.
References
- Han, P.; Ivanovski, S. Saliva—Friend and Foe in the COVID-19 Outbreak. Diagnostics 2020, 10, 290.
- Sabino-Silva, R.; Jardim, A.C.G.; Siqueira, W.L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin. Oral Investig. 2020, 24, 1–3.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- Zupin, L.; Pascolo, L.; Crovella, S. Is FURIN gene expression in salivary glands related to SARS-CoV-2 infectivity through saliva? J. Clin. Pathol. 2020, 74, 209–211.
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a reliable tool to detect Sars-CoV-2. J. Infect. 2020, 81, e45–e50.
- Azzi, L.; Carcano, G.; Dalla Gasperina, D.; Sessa, F.; Maurino, V.; Baj, A. Two cases of COVID-19 with positive salivary and negative pharyngeal or respiratory swabs at hospital discharge: A rising concern. Oral Dis. 2020, 27, 707–709.
- Chen, L.; Zhao, J.; Peng, J.; Li, X.; Deng, X.; Geng, Z.; Shen, Z.; Guo, F.; Zhang, Q.; Jin, Y.; et al. Detection of 2019-nCoV in Saliva and Characterization of Oral Symptoms in COVID-19 Patients. SSRN Electron. J. 2020, 53, e12923.
- Mi Seon, H.; Moon-Woo, S.; Eun Young, H.; Ji Hong, P.; Namhee, K.; Sue, S.; Sung Im, C.; Sung Sup, P.; Eun Hwa, C. Sequential Analysis of Viral Load in a Neonate and Her Mother Infected With Severe Acute Respiratory Syndrome Coronavirus 2|Clinical Infectious Diseases|Oxford Academic. Clin. Infect. Dis. 2020, 71, 2236–2239.
- To, K.K.W.; Tsang, O.T.Y.; Yip, C.C.Y.; Chan, K.H.; Wu, T.C.; Chan, J.M.C.; Leung, W.S.; Chik, T.S.H.; Choi, C.Y.C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva | Clinical Infectious Diseases|Oxford Academic. Clin. Infect. Dis. 2020, 71, 841–843.
- To, K.K.W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.Y.; Cai, J.P.; Chan, J.M.C.; Chik, T.S.H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574.
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B.F.; et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv 2020, 383, 1283–1286.
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369.
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389.
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease-2019 (COVID-19): A cross-sectional study. Clin. Microbiol. Infect. 2020, 27, 285.e1–285.e4.
- Sirikhetkon, S.; Shrestha, M.; Okada, P.A.; Prasert, K. Diagnostic Accuracy of Saliva for SARS-CoV-2 Detection in State-sponsored Quarantine in Thailand. Outbreak Surveill. Investig. Response J. 2021, 14, 12–19.
- Basso, D.; Aita, A.; Padoan, A.; Cosma, C.; Navaglia, F.; Moz, S.; Contran, N.; Zambon, C.F.; Maria Cattelan, A.; Plebani, M. Salivary SARS-CoV-2 antigen rapid detection: A prospective cohort study. Clin. Chim. Acta 2021, 517, 54–59.
- To, K.K.; Tsang, O.T.Y.; Chik-Yan Yip, C. Consistent detection of novel coronavirus in saliva. Clin. Infect. Dis 2019, 71, 841–843.
- Azzi, L.; Baj, A.; Alberio, T.; Lualdi, M.; Veronesi, G.; Carcano, G.; Ageno, W.; Gambarini, C.; Maffioli, L.; Di Saverio, S. Rapid Salivary Test suitable for a mass screening program to detect SARS-CoV-2: A diagnostic accuracy study. J. Infect. 2020, 81, e75–e78.
- Cheng, V.C.C.; Wong, S.-C.; Chen, J.H.K.; Yip, C.C.Y.; Chuang, V.W.M.; Tsang, O.T.Y.; Sridhar, S.; Chan, J.F.W.; Ho, P.-L.; Yuen, K.-Y. Escalating infection control response to the rapidly evolving epidemiology of the Coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020, 41, 493–498.
- Han, M.S.; Seong, M.-W.; Kim, N.; Shin, S.; Cho, S.I.; Park, H.; Kim, T.S.; Park, S.S.; Choi, E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020, 26, 2497–2499.
- Han, M.S.; Seong, M.-W.; Heo, E.Y.; Park, J.H.; Kim, N.; Shin, S.; Cho, S.I.; Park, S.S.; Choi, E.H. Sequential analysis of viral load in a neonate and her mother infected with SARS-CoV-2. Clin. Infect. Dis. 2020, 71, 2236–2239.
- Iwasaki, S.; Fujisawa, S.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Fukumoto, T.; Sato, K.; Oguri, S.; Taki, K.; Senjo, H. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020, 81, e145–e147.
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020, 71, 841–843.
- Yoon, J.G.; Yoon, J.; Song, J.Y.; Yoon, S.-Y.; Lim, C.S.; Seong, H.; Noh, J.Y.; Cheong, H.J.; Kim, W.J. Clinical Significance of a High SARS-CoV-2 Viral Load in the Saliva. J. Korean Med. Sci. 2020, 35, e195.
- Zhu, J.; Guo, J.; Xu, Y.; Chen, X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J. Infect. 2020, 81, e48–e50.
- Jamal, A.J.; Mohammad, M.; Coomes, E.; Powis, J.; Li, A.; Paterson, A.; Anceva-Sami, S.; Barati, S.; Crowl, G.; Faheem, A. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). medRxiv 2020, 72, 1064–1066.
- Leung, E.C.; Chow, V.C.; Lee, M.K.; Lai, R.W. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J. Med. Virol. 2020, 93, 533–536.
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M. Clinical evaluation of self-collected saliva by RT-qPCR, direct RT-qPCR, RT-LAMP, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20.
- Tajima, Y.; Suda, Y.; Yano, K. A case report of SARS-CoV-2 confirmed in saliva specimens up to 37 days after onset: Proposal of saliva specimens for COVID-19 diagnosis and virus monitoring. J. Infect. Chemother. 2020, 26, 1086–1089.
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; Williamson, D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00776-20.
- Otto, M.P.; Darles, C.; Valero, E.; Benner, P.; Dutasta, F.; Janvier, F. Posterior oropharyngeal salivafor the detection of SARS-CoV-2. Clin. Infect. Dis. 2020, 71, 2939–2946.
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.; Slowey, P.; Almas, K. Human Saliva Collection Devices for Proteomics: An Update. Int. J. Mol. Sci. 2016, 17, 846.
- Khurshid, Z. Salivary point-of-care technology. Eur. J. Dent. 2018, 12, 1–2.
- FDA. U.S. Food and Drug Administration Emergency Use Authorizations; FDA: Muntinlupa, PH, USA, 2021. Available online: (accessed on 26 March 2021).
- Sullivan, P.S.; Sailey, C.; Guest, J.L.; Guarner, J.; Siegler, A.J.; Valentine-Graves, M.; Gravens, L.; del Rio, C.; Sanchez, T.H. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: Protocol to validate the sufficiency of provider-observed home-collected blood, saliva and oropharyngeal samples (Preprint). JMIR Public Health Surveill. 2020, 6, e19054.
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 1–5.
- Fulgent Therapeutics, L. Fulgent COVID-19 by RT-PCR test EUA Summary 2 Device Description and Test Principle. Available online: (accessed on 23 July 2020).
