SARS-CoV-2 nucleocapsid protein (NCoV2) plays a key role in various processes related to the viral replication cycle such as the RNA genome packaging, interaction with other viral proteins. It has thus been a target for new drug development.
- coronavirus nucleocapsid protein
- molecular flexibility
- protein structure
- protein dynamics
- liquid-liquid phase separation
- neutron scattering
1. Introduction
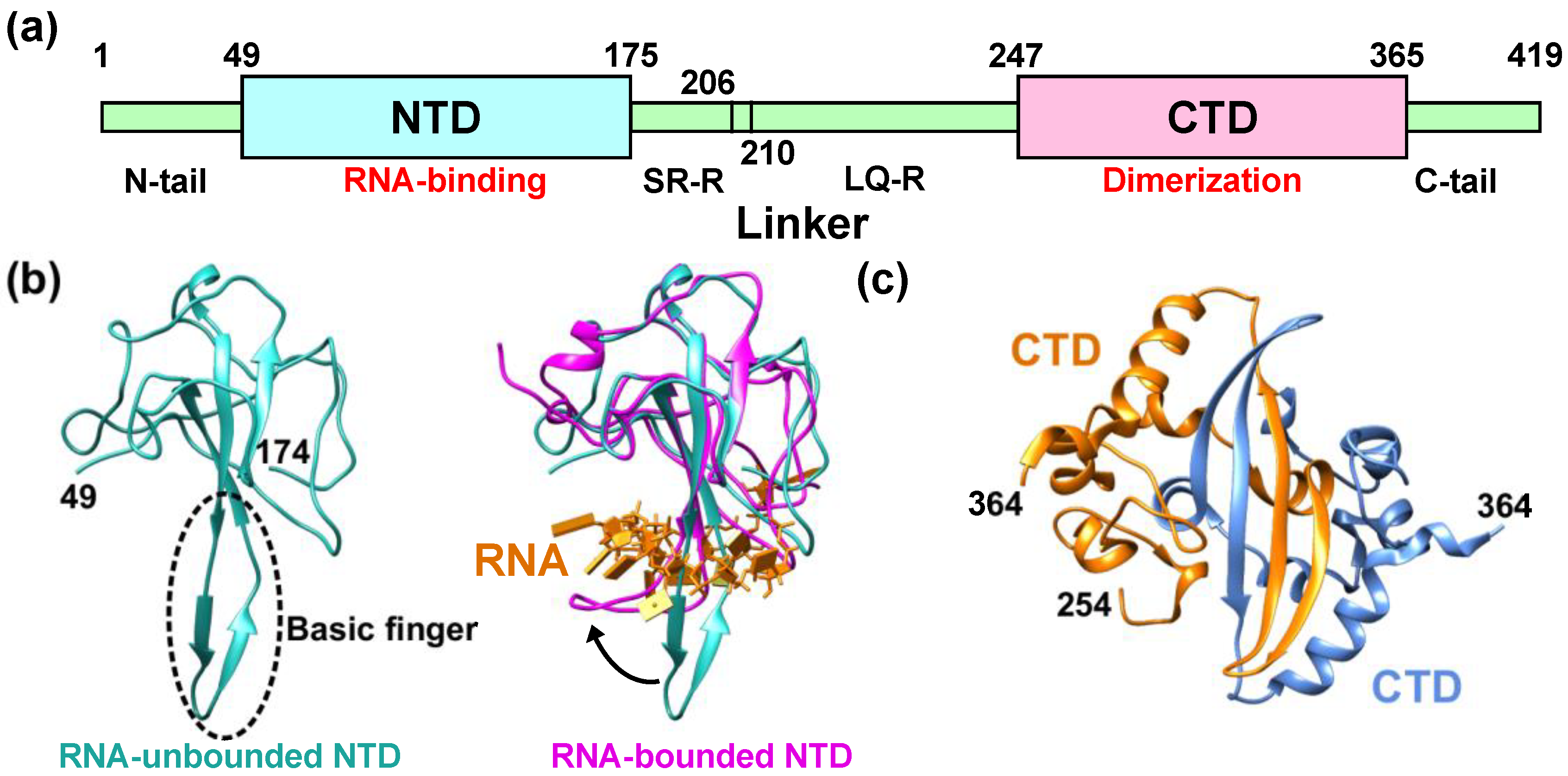 Figure 1. Structural features of the nucleocapsid protein of SARS-CoV-2 (NCoV2). (a) Schematic of NCoV2, which consists of two domains, i.e., the N-terminal domain (NTD) and the C-terminal domain (CTD); (b) atomic structure of the NTD in the RNA-unbounded form (left; PDB ID: 6M3M [12]) and the single-stranded RNA-bounded form (right; PDB ID: 7ACT [13]) are shown in light sea green and magenta, respectively. In the right panel of (b), RNA is denoted in orange and the RNA-unbounded form of the NTD is superimposed for structural comparison. The direction of the movement of the basic finger upon RNA-binding is shown by a black arrow; (c) CTD dimer interface. The CTD monomers (PDB ID: 6WZO [15]) are shown in orange and marine blue, respectively. In (b,c), the residue numbers at both termini of the structures that are visible in the crystal structures are denoted. All the models are depicted using UCSF Chimera [16].
Figure 1. Structural features of the nucleocapsid protein of SARS-CoV-2 (NCoV2). (a) Schematic of NCoV2, which consists of two domains, i.e., the N-terminal domain (NTD) and the C-terminal domain (CTD); (b) atomic structure of the NTD in the RNA-unbounded form (left; PDB ID: 6M3M [12]) and the single-stranded RNA-bounded form (right; PDB ID: 7ACT [13]) are shown in light sea green and magenta, respectively. In the right panel of (b), RNA is denoted in orange and the RNA-unbounded form of the NTD is superimposed for structural comparison. The direction of the movement of the basic finger upon RNA-binding is shown by a black arrow; (c) CTD dimer interface. The CTD monomers (PDB ID: 6WZO [15]) are shown in orange and marine blue, respectively. In (b,c), the residue numbers at both termini of the structures that are visible in the crystal structures are denoted. All the models are depicted using UCSF Chimera [16].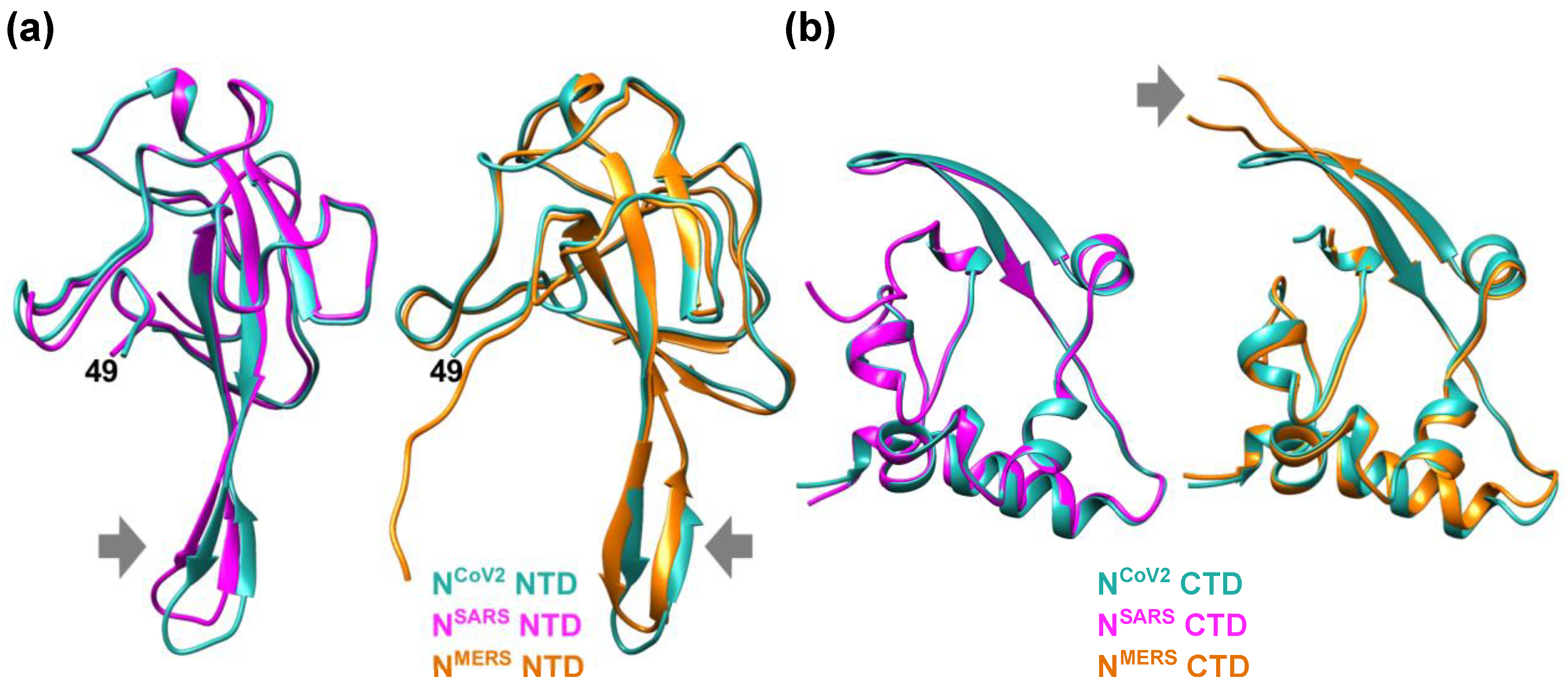 Figure 2. Comparison of the NTD and CTD structures of the nucleocapsid protein of different coronaviruses. (a) (left) Comparison of the NTD structures of the nucleocapsid protein of SARS-CoV-2 (NCoV2; PDB ID: 6M3M [12]) and SARS-CoV (NSARS; PDB ID: 2OFZ [21]), which are shown in marine blue and magenta, respectively. (right) Comparison of NCoV2 NTD and the NTD of the nucleocapsid protein of MERS-CoV (NMERS; PDB ID: 4UD1 [22]) shown in orange. Note that NCoV2 NTD in the left and right panels are displayed at slightly different angles to focus on the structural differences from the NSARS and NMERS counterparts; (b) (left) Comparison of the CTD structures of NCoV2 (PDB ID: 7C22 [18]) and of NSARS (PDB ID: 2CJR [23]), which are shown in marine blue and magenta, respectively. (right) Comparison of NCoV2 CTD and the CTD structure of NMERS (PDB ID: 6G13 [24]) shown in orange. In both (a,b), the major structural differences are shown by grey arrows. All the models are depicted using UCSF Chimera [16].
Figure 2. Comparison of the NTD and CTD structures of the nucleocapsid protein of different coronaviruses. (a) (left) Comparison of the NTD structures of the nucleocapsid protein of SARS-CoV-2 (NCoV2; PDB ID: 6M3M [12]) and SARS-CoV (NSARS; PDB ID: 2OFZ [21]), which are shown in marine blue and magenta, respectively. (right) Comparison of NCoV2 NTD and the NTD of the nucleocapsid protein of MERS-CoV (NMERS; PDB ID: 4UD1 [22]) shown in orange. Note that NCoV2 NTD in the left and right panels are displayed at slightly different angles to focus on the structural differences from the NSARS and NMERS counterparts; (b) (left) Comparison of the CTD structures of NCoV2 (PDB ID: 7C22 [18]) and of NSARS (PDB ID: 2CJR [23]), which are shown in marine blue and magenta, respectively. (right) Comparison of NCoV2 CTD and the CTD structure of NMERS (PDB ID: 6G13 [24]) shown in orange. In both (a,b), the major structural differences are shown by grey arrows. All the models are depicted using UCSF Chimera [16].
2. Perspective–Toward Structural and Dynamical Characterization of NCoV2 Droplets
The recent findings that NCoV2, RNA, other viral proteins and a stress granule protein are concentrated into a different phase from its surroundings [25][26][27][28][29][30][31] suggest that the NCoV2 droplet formed through liquid-liquid phase separation (LLPS) is the main arena where molecular events critical for the virus replication take place. It is thus indispensable to reveal the internal organization of NCoV2 droplets, especially the structure of NCoV2 in droplets formed with various binding partners to develop a new intervention technique targeting NCoV2 as well as to understand the molecular mechanism of biological functions of NCoV2 in detail. Furthermore, the molecules within the droplets are not frozen to the spot but fluctuate dynamically around their average conformation under the influence of thermal energy. The nature of these structural fluctuations is closely related to the interaction, the stability and the function of the proteins. Thus, unraveling the structure and structural fluctuation of NCoV2 in the droplets formed in various conditions is essential and the research in this direction would occupy an important position in this research field. A powerful method to characterize the molecular structure of NCoV2
in droplets would be small-angle neutron scattering (SANS). This technique has been used to characterize the structure of viral proteins in isolated state and in complex with other viral components . However, to the author’s knowledge, the structural and dynamical analysis of droplets formed by virus-related proteins has not been carried out so far. Even though there are excellent reviews and books on neutron scattering , some basic information on neutron scattering is given below in order to describe a possible method to reveal the structure and dynamics of NCoV2
droplets. In neutron scattering, the strength of scattering (coherent scattering length) depends on the type of atoms bombarded by neutron. The major feature of coherent neutron scattering is that the scattering length of hydrogen and its isotope deuterium is largely different and thus deuteration technique plays a significant role in the structure analysis of a complex system that consists of different kinds of components . In the case of the solution samples such as protein solutions, the scattering signal in SANS measurements derives from the difference (contrast) in the scattering length density (SLD: the total scattering length of a molecule divided by its volume) between solutes and solvents. Figure 3a shows the variations of the SLD of major biomolecules (proteins, lipids, DNA and RNA) as a function of the fraction of D2
O in the solvent. As the D2
O content in the solvent increases, the labile hydrogen atoms in the solutes are exchanged for deuterium atoms, which modifies the SLD of the solutes. The SLD of perdeuterated proteins (denoted as “D-protein” in Figure 3a) is much larger than that of the hydrogenated counterparts because all the hydrogen atoms including those in methyl and ethyl groups are replaced with deuterium atoms. As seen in Figure 3a, the SLD of the solvent containing 40% D2
O is equal to that of the hydrogenated protein (H-protein), meaning that the scattering contrast of the H-protein is zero. In this condition, the contribution of the H-protein to the scattering signal is effectively zero and hence it is “invisible” to neutron. Even though techniques to perdeuterate or partially deuterate RNA molecules by chemical synthesis, in vitro transcription and in vivo production have been established for NMR studies , production of the sufficient amount of perdeuterated RNA for neutron experiments is more expensive and/or more difficult than production of perdeuterated protein and thus there have been only a very few SANS studies using perdeuterated RNA[38][46]. Therefore, in the following, only RNA in the hydrogenated state (H-RNA) is considered.
. Therefore, in the following, only RNA in the hydrogenated state (H-RNA) is considered.Figure
Figure 3c illustrates a promising method to characterize the NCoV2 structure in droplets. In the case where droplets are formed by NCoV2, RNA and another protein (Figure 3c), three kinds of samples would be required to obtain the scattering signal from each protein: (1) droplets formed by H-NCoV2, H-RNA and the hydrogenated another protein (H-AP), (2) those formed by H-NCoV2, H-RNA and the perdeuterated another protein (D-AP) and (3) those formed by the perdeuterated NCoV2 (D-NCoV2), H-RNA and D-AP. The SANS measurements in these samples in the ~65% D2O solvent, the SLD of which matches that of H-RNA, provide three kinds of scattering data where each protein contribution is different. From these data, the scattering data arising from each protein can be extracted, leading to the elucidation of its conformation together with how each protein is arranged inside the droplets.
The structural fluctuation of the molecules in the droplets are obtained by incoherent neutron scattering (iNS). This technique measures the intensity of neutron scattered by the samples as a function of the momentum and energy change of the neutron, from which the amplitude, the frequency and their distribution of atomic motions at pico- to nanosecond timescale at ångström length scale are estimated. The values of incoherent scattering cross-section of atoms found in biomolecules are shown in Figure 3b. It is found that the scattering cross-section of hydrogen atoms is much larger than any other atom and deuterium, meaning that iNS provides information of the motions of hydrogen atoms. Furthermore, the measurements in 100% D2O buffer can minimize the solvent contribution to the iNS spectra. Since the hydrogen atoms are quasi-uniformly distributed throughout the molecules in the biological systems, the dynamical information averaged over the whole molecule is obtained. In the case of proteins, the observed motions reflect the fluctuations of amino acid side chains and backbones because hydrogen atoms are bound to these chemical groups. The schematic illustration of the dynamical analysis of the NCoV2 droplets is depicted in Figure 3d. For droplets generated from NCoV2, RNA and another protein, two types of samples are required: (1) droplets formed by H-NCoV2, H-RNA and D-AP, (2) those formed by D-NCoV2, H-RNA and D-AP. By subtracting the iNS spectra of the latter from those of the former measured in 100% D2O, the remaining spectra arise from the structural fluctuation of the NCoV2 molecules, thereby providing dynamical information on NCoV2 inside the droplets. Instead, the dynamics of another protein in the droplets can be extracted in a similar manner.
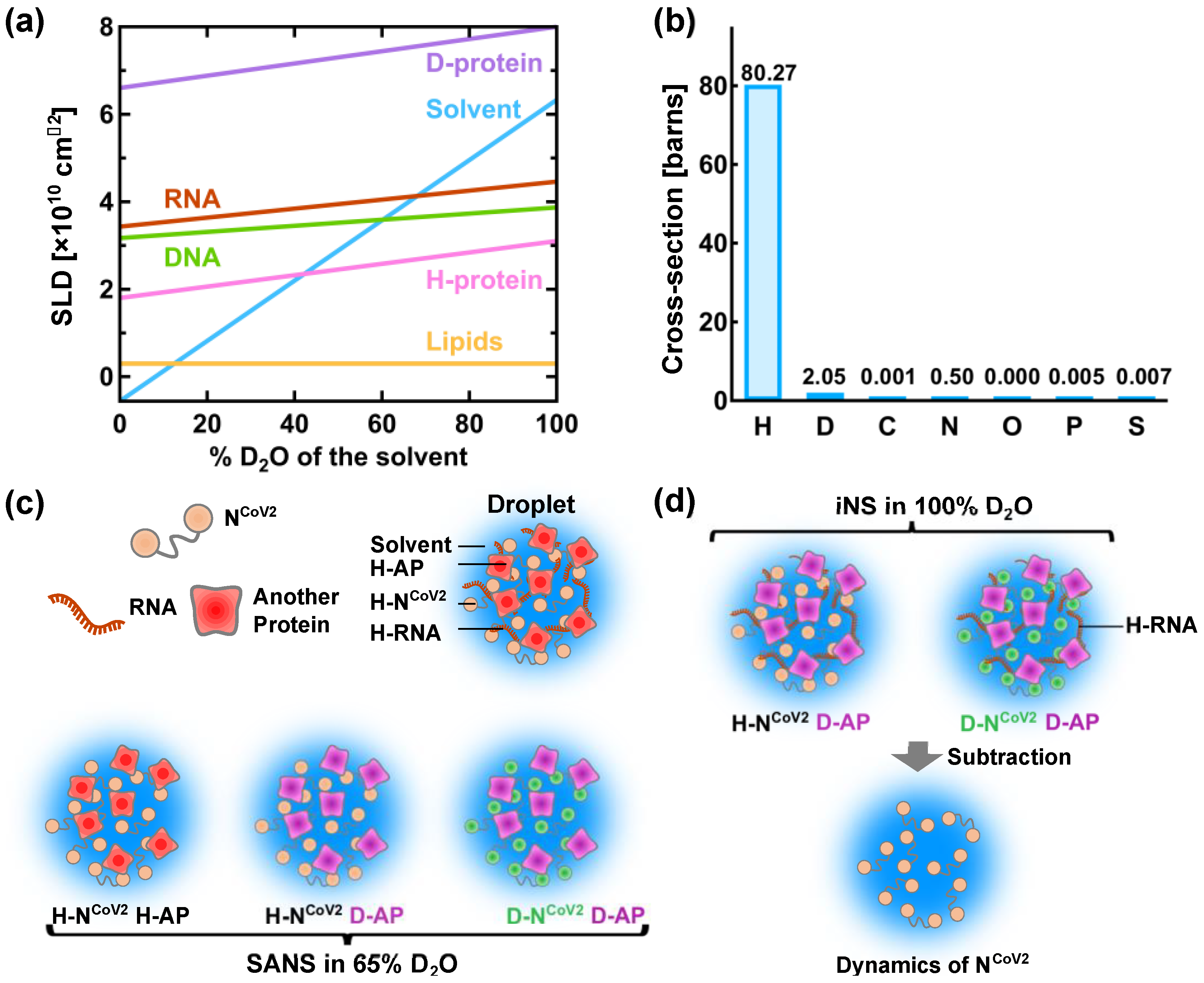 Figure 3. A promising method for physical characterization of the NCoV2 droplets using neutron scattering. (a) Variation of the neutron scattering length density (SLD) of biomacromolecules as a function of the heavy water concentration in the solvent. H-protein and D-protein denote the hydrogenated and perdeuterated proteins, respectively. The values of the SLD of H-protein, D-protein, lipids and solvent are taken from [37], and those of DNA and RNA are taken from [39]; (b) values of incoherent neutron scattering cross-section of atoms found in biomolecules and an isotope of hydrogen atom, deuterium. Note that 1 barn = 10−24 cm2. The values are taken from [47]; (c) schematic illustration of the structural analysis of the NCoV2 droplets using contrast-matching small-angle neutron scattering (SANS). The components that are “invisible” to neutron are shown in the same color as solvent. The prefixes H- and D- denote “hydrogenated” and “perdeuterated”, respectively, which are shown in different colors. AP denotes another protein; (d) schematic illustration of the molecular dynamics analysis of NCoV2 droplets using incoherent neutron scattering (iNS) combined with deuteration technique. For detailed explanations of (c,d), refer to the main text.
Figure 3. A promising method for physical characterization of the NCoV2 droplets using neutron scattering. (a) Variation of the neutron scattering length density (SLD) of biomacromolecules as a function of the heavy water concentration in the solvent. H-protein and D-protein denote the hydrogenated and perdeuterated proteins, respectively. The values of the SLD of H-protein, D-protein, lipids and solvent are taken from [37], and those of DNA and RNA are taken from [39]; (b) values of incoherent neutron scattering cross-section of atoms found in biomolecules and an isotope of hydrogen atom, deuterium. Note that 1 barn = 10−24 cm2. The values are taken from [47]; (c) schematic illustration of the structural analysis of the NCoV2 droplets using contrast-matching small-angle neutron scattering (SANS). The components that are “invisible” to neutron are shown in the same color as solvent. The prefixes H- and D- denote “hydrogenated” and “perdeuterated”, respectively, which are shown in different colors. AP denotes another protein; (d) schematic illustration of the molecular dynamics analysis of NCoV2 droplets using incoherent neutron scattering (iNS) combined with deuteration technique. For detailed explanations of (c,d), refer to the main text.3. Conclusions
References
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502.
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885.
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 131.
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208.
- Astuti, I. Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412.
- Surjit, M.; Lal, S.K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evol. 2008, 8, 397–405.
- Chang, C.-K.; Hou, M.-H.; Chang, C.-F.; Hsiao, C.-D.; Huang, T.-H. The SARS coronavirus nucleocapsid protein—Forms and functions. Antivir. Res. 2014, 103, 39–50.
- Zhu, G.; Zhu, C.; Zhu, Y.; Sun, F. Minireview of progress in the structural study of SARS-CoV-2 proteins. Curr. Res. Microb. Sci. 2020, 1, 53–61.
- Chang, C.-K.; Chen, C.-M.M.; Chiang, M.-H.; Hsu, Y.-L.; Huang, T.-H. Transient Oligomerization of the SARS-CoV N Protein—Implication for Virus Ribonucleoprotein Packaging. PLoS ONE 2013, 8, e65045.
- Katsuma, S.; Kokusho, R. A Conserved Glycine Residue Is Required for Proper Functioning of a Baculovirus VP39 Protein. J. Virol. 2017, 91, e02253-16.
- Kang, S.; Yang, M.; Hong, Z.; Zhang, L.; Huang, Z.; Chen, X.; He, S.; Zhou, Z.; Zhou, Z.; Chen, Q.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020, 10, 1228–1238.
- Dinesh, D.C.; Chalupska, D.; Silhan, J.; Koutna, E.; Nencka, R.; Veverka, V.; Boura, E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020, 16, e1009100.
- Tan, Y.W.; Fang, S.; Fan, H.; Lescar, J.; Liu, D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006, 34, 4816–4825.
- Ye, Q.; West, A.M.V.; Silletti, S.; Corbett, K.D. Architecture and self-assembly of the SARS-CoV -2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612.
- Zinzula, L.; Basquin, J.; Bohn, S.; Beck, F.; Klumpe, S.; Pfeifer, G.; Nagy, I.; Bracher, A.; Hartl, F.U.; Baumeister, W. High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 2021, 538, 54–62.
- Zhou, R.; Zeng, R.; Von Brunn, A.; Lei, J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol. Biomed. 2020, 1, 1–11.
- Takeda, M.; Chang, C.-K.; Ikeya, T.; Güntert, P.; Chang, Y.-H.; Hsu, Y.-L.; Huang, T.-H.; Kainosho, M. Solution Structure of the C-terminal Dimerization Domain of SARS Coronavirus Nucleocapsid Protein Solved by the SAIL-NMR Method. J. Mol. Biol. 2008, 380, 608–622.
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623.
- Saikatendu, K.S.; Joseph, J.S.; Subramanian, V.; Neuman, B.W.; Buchmeier, M.J.; Stevens, R.C.; Kuhn, P. Ribonucleocapsid Formation of Severe Acute Respiratory Syndrome Coronavirus through Molecular Action of the N-Terminal Domain of N Protein. J. Virol. 2007, 81, 3913–3921.
- Papageorgiou, N.; Lichière, J.; Baklouti, A.; Ferron, F.; Sévajol, M.; Canard, B.; Coutard, B. Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 192–202.
- Chen, C.-Y.; Chang, C.-K.; Chang, Y.-W.; Sue, S.-C.; Bai, H.-I.; Riang, L.; Hsiao, C.-D.; Huang, T.-H. Structure of the SARS Coronavirus Nucleocapsid Protein RNA-binding Dimerization Domain Suggests a Mechanism for Helical Packaging of Viral RNA. J. Mol. Biol. 2007, 368, 1075–1086.
- Van Nguyen, T.H.; Lichière, J.; Canard, B.; Papageorgiou, N.; Attoumani, S.; Ferron, F.; Coutard, B. Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 8–15.
- Jasmine Cubuk; Jhullian J. Alston; J. Jeremías Incicco; Sukrit Singh; Melissa D. Stuchell-Brereton; Michael D. Ward; Maxwell I. Zimmerman; Neha Vithani; Daniel Griffith; Jason A. Wagoner; et al.Gregory R. BowmanKathleen B. HallAndrea SorannoAlex S. Holehouse The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nature Communications 2021, 12, 1-17, 10.1038/s41467-021-21953-3.
- Hui Chen; Yang Cui; Xuling Han; Wei Hu; Min Sun; Yong Zhang; Pei-Hui Wang; Guangtao Song; Wei Chen; Jizhong Lou; et al. Liquid–liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Research 2020, 30, 1143-1145, 10.1038/s41422-020-00408-2.
- Adriana Savastano; Alain Ibáñez De Opakua; Marija Rankovic; Markus Zweckstetter; Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nature Communications 2020, 11, 1-10, 10.1038/s41467-020-19843-1.
- Theodora Myrto Perdikari; Anastasia C. Murthy; Veronica H. Ryan; Scott Watters; Mandar T. Naik; Nicolas L. Fawzi; SARS‐CoV‐2 nucleocapsid protein phase‐separates with RNA and with human hnRNPs. The EMBO Journal 2020, 39, e106478, 10.15252/embj.2020106478.
- Shan Lu; Qiaozhen Ye; Digvijay Singh; Yong Cao; Jolene K. Diedrich; John R. Yates; Elizabeth Villa; Don W. Cleveland; Kevin D. Corbett; The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nature Communications 2021, 12, 1-15, 10.1038/s41467-020-20768-y.
- Christopher R. Carlson; Jonathan B. Asfaha; Chloe M. Ghent; Conor J. Howard; Nairi Hartooni; Maliheh Safari; Alan D. Frankel; David O. Morgan; Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Molecular Cell 2020, 80, 1092-1103.e4, 10.1016/j.molcel.2020.11.025.
- Christiane Iserman; Christine A. Roden; Mark A. Boerneke; Rachel S.G. Sealfon; Grace A. McLaughlin; Irwin Jungreis; Ethan J. Fritch; Yixuan J. Hou; Joanne Ekena; Chase A. Weidmann; et al.Chandra L. TheesfeldManolis KellisOlga G. TroyanskayaRalph S. BaricTimothy P. SheahanKevin M. WeeksAmy S. Gladfelter Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Molecular Cell 2020, 80, 1078-1091.e6, 10.1016/j.molcel.2020.11.041.
- Jensen, M.R.; Communie, G.; Ribeiro, E.A.; Martinez, N.; Desfosses, A.; Salmon, L.; Mollica, L.; Gabel, F.; Jamin, M.; Longhi, S.; et al. Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. USA 2011, 108, 9839–9844.
- Blanchard, L.; Tarbouriech, N.; Blackledge, M.; Timmins, P.; Burmeister, W.P.; Ruigrok, R.W.; Marion, D. Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 2004, 319, 201–211.
- He, L.; Piper, A.; Meilleur, F.; Hernandez, R.; Heller, W.T.; Brown, D.T. Conformational Changes in Sindbis Virus Induced by Decreased pH Are Revealed by Small-Angle Neutron Scattering. J. Virol. 2012, 86, 1982–1987.
- Das, N.C.; Warren, G.T.; Kao, C.C.; Dragnea, B.G.; Ni, P.; Sokol, P.E. Small Angle Scattering Study of the Structure and Organization of RNAs and Protein of Brome Mosaic Virus (BMV) Capsid Protein. Phys. Procedia 2014, 60, 101–109.
- He, L.; Piper, A.; Meilleur, F.; Myles, D.A.A.; Hernández, R.; Brown, D.T.; Heller, W.T. The Structure of Sindbis Virus Produced from Vertebrate and Invertebrate Hosts as Determined by Small-Angle Neutron Scattering. J. Virol. 2010, 84, 5270–5276.
- Svergun, D.I.; Koch, M.H.; Timmins, P.A.; May, R.P. Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules; Oxford University Press: Oxford, UK, 2013.
- Mahieu, E.; Gabel, F. Biological small-angle neutron scattering: Recent results and development. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 715–726.
- Fitter, J.; Gutberlet, T.; Katsaras, J. (Eds.) Neutron Scattering in Biology; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 978-3-540-29108-4.
- Heller, W.T. Small-angle neutron scattering and contrast variation: A powerful combination for studying biological structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 1213–1217.
- Gabel, F. Small-Angle Neutron Scattering for Structural Biology of Protein–RNA Complexes. Methods Enzymol. 2015, 558, 391–415.
- Mirandela, G.D.; Tamburrino, G.; Ivanović, M.T.; Strnad, F.M.; Byron, O.; Rasmussen, T.; Hoskisson, P.A.; Hub, J.S.; Zachariae, U.; Gabel, F.; et al. Merging In-Solution X-ray and Neutron Scattering Data Allows Fine Structural Analysis of Membrane–Protein Detergent Complexes. J. Phys. Chem. Lett. 2018, 9, 3910–3914.
- Lapinaite, A.; Carlomagno, T.; Gabel, F. Small-Angle Neutron Scattering of RNA–Protein Complexes. Methods Mol. Biol. 2020, 2113, 165–188.
- Lu, K.; Miyazaki, Y.; Summers, M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 2010, 46, 113–125.
- Duss, O.; Lukavsky, P.J.; Allain, F.H.-T. Isotope Labeling and Segmental Labeling of Larger RNAs for NMR Structural Studies BT—Isotope labeling in Biomolecular NMR. In Isotope Labeling in Biomolecular NMR.; Atreya, H.S., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 121–144. ISBN 978-94-007-4954-2.
- Lapinaite, A.; Simon, B.; Skjaerven, L.; Rakwalska-Bange, M.; Gabel, F.; Carlomagno, T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nat. Cell Biol. 2013, 502, 519–523.
- Sears, V.F. Neutron scattering lengths and cross sections. Neutron News 1992, 3, 26–37.
