Mortality rates of mucormycosis can reach up to 90%, due to the mucoralean antifungal drug resistance and the lack of effective therapies. Non-canonical RNAi pathway (NCRIP) regulates the expression of mRNAs by degrading them in a specific manner. Its mechanism binds dsRNA but only cuts ssRNA. NCRIP exhibits a diversity of functional roles. It represses the epimutational pathway and the lack of NCRIP increases the generation of drug resistant strains. NCRIP also regulates the control of retrotransposons expression, playing an essential role in genome stability.
- virulence
- antifungal resistance
- non-canonical RNAi
- epimutant
- R3B2
- RdRP
- transposon
- genome stability
- Mucorales
- mucormycosis
1. Introduction
The innate antifungal drug resistance and virulence observed in Mucorales must be based on specific mechanisms not found in other fungi [1][2][3][4,19,20]. Among these mechanisms, the most studied and related to virulence and antifungal drug resistance is the RNAi mechanism. The first RNAi-based mechanism discovered in Mucorales was the exogenously induced RNAi pathway, a defense system against transgenes and plasmids. Later, two endogenous RNAi pathways were discovered: the canonical and the non-canonical pathways (Figure 1) [4][5][6][7][8][9][15,21,22,23,24,25]. A subtype of the canonical pathway, the epimutational pathway, is directly involved in the generation of transient antifungal drug resistances. This pathway is capable of silencing the gene that encodes the target proteins inhibited by the antifungal compound [2][19]. In the absence of the target protein, the antifungal compound is unable of inhibiting the regular growth of the fungus. The non-canonical RNA interference pathway (NCRIP) was initially described as a non-specific RNA degradation mechanism, although further studies demonstrated its role in regulating several processes [7][10][11][23,26,27].
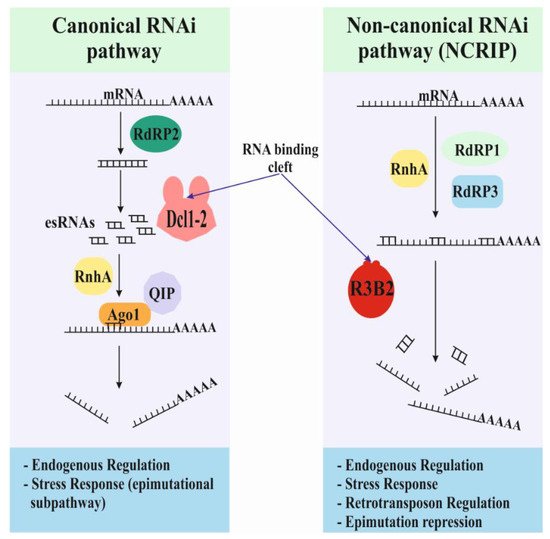
Figure 1. Models and functions for the two endogenous RNAi-based pathways. There are two endogenous RNAi-based pathways in Mucorales, the canonical and non-canonical pathways. The canonical pathway uses the conserved core machinery based on the activities of Dicer and Argonaute proteins. The non-canonical pathway relies on a new RNase type III: the R3B2 protein. R3B2 binds double-strand RNA (dsRNA) but only cuts single-strand RNA (ssRNA).
2. Structural and Functional Analysis of R3B2
The characterization of NCRIP and its degradation products in the Dicer null mutants led to the discovery of the atypical RNase R3B2 in M. lusitanicus [7][23]. This RNase shows an RNase III-like domain (RIIID) together with two double-strand RNA binding domains (dsRBD), which is an unusual feature for a protein with a hypothesized activity against single-stranded RNA (ssRNA). In fact, both analyses in vivo by the yeast two-hybrid interaction assay and in vitro by size exclusion chromatography and static light scattering indicate that R3B2 dimerizes through the RIIID [12][44]. Likewise, bacterial RNase III and the two RIIIDs of eukaryotic Dicer and Drosha type RNases III dimerizes to cut each strand in the double-stranded RNA molecule (dsRNA) [13][14][15][45,46,47]. The apparent contradiction between the substrate preference of RNase III proteins and the ssRNA degradations products observed in the NCRIP supposed the basis for the thorough study of this new family of RNase III proteins related to R3B2.
The recent purification of R3B2 protein of M. lusitanicus revealed that the RNase uses both dsRBDs to bind either ssRNA or dsRNA in a sequence-independent manner [12][44]. These results suggest an additional protein that can interact with R3B2 and determine the targeted mRNAs regulated by the NCRIP. Interactions with RNase III proteins to manage or modulate their function are observed from plants [16][48] to animals [17][35].
3. Interaction between NCRIP and the Canonical RNAi Pathways: Regulation of the Epimutational Antifungal Drug Resistance
The NCRIP negatively regulates the generation of antifungal drug resistant epimutants through its control over the canonical RNAi mechanism [10][26]. The endogenous canonical pathway constitutively produces a particular class of esRNAs, the ex-siRNAs, which regulate hundreds of target mRNAs [6][22]. Recently, a subpathway of this endogenous canonical RNAi mechanism has been discovered: the epimutational pathway [2][19]. This epimutational pathway produces specific ex-siRNAs in response to an external stimulus. The specific ex-siRNAs are produced only from the key target genes involved in response to the external stimulus. The silencing of these target genes implies a transient and unstable adaptation to the new external conditions. The specific ex-siRNAs are mitotically passed on to the descendant epimutants as long as the external conditions are maintained. The silencing of the target genes and the production of ex-siRNAs stop when the external stimulus disappears [2][19].
4. Role of NCRIP in the Integrity and Defense of the Genome
Despite NCRIP negatively regulates the epimutational pathway, its most specific component, R3B2, is also necessary for the transgene-induced RNAi silencing. The role of R3B2 in the transgene-induced RNAi pathway was confirmed after transforming r3b2 mutants with different exogenous triggering molecules. The frequency of silenced transformant was reduced in the r3b2 mutants, indicating that R3B2 is critical for efficient silencing of exogenous molecules [7][23]. The minor role of R3B2 in the canonical RNAi pathways is likely related to a putative interaction of the Dicer proteins, although its function in these pathways is still unknown [18][60]. The discrimination of R3B2 between the canonical and non-canonical pathways is also undetermined. The proposed hypothesis suggests a likely collaboration of R3B2 with either Dicer or the RdRP proteins to participate in the canonical RNAi mechanism or NCRIP, respectively. Results indicated a negative regulatory role of NCRIP over the canonical repression of transposons, similar to the repression of NCRIP over the epimutational RNAi pathway.
5. The NCRIP Mechanism Regulates Virulence in Mucorales
The lack of an active NCRIP mechanism results in misregulation of the genetic response of mucoralean spores during the confrontation with macrophages, the first stage of the infection [11][19][27,65]. The studies confronting the sRNAs produced explicitly by the NCRIP mechanism with the genome sequence of M. lusitanicus found that this pathway regulates many genes [7][23]. Therefore, it is expected a pleiotropic role of NCRIP in the fungal physiology of Mucorales, controlling diverse processes. In fact, the phenotypic analysis of strains lacking the NCRIP activity confirmed the role of this mechanism in complex cellular processes such as the response to oxidative stress and the development of zygospores during sexual development [7][23]. The phenotype affecting the production of zygospores can be related to the regulatory role of NCRIP over the canonical RNAi pathway during the control of the centromeric transposons. In Cryptococcus neoformans, RNAi-related mechanisms protect the genome from transposon activity during mating, securing the genomic integrity of the progeny [20][66]. The second phenotype, the misregulated response to oxidative stress, might be related to a functional role of NCRIP in a natural scenario where this stress is activated, for instance, during the oxidative attack of macrophages after phagocytosis. Thus, thorough studies investigated the role of NCRIP during the interaction of macrophage and mucoralean spores. These reports found that the genetic response of M. lusitanicus during the interaction with macrophages showed hundreds of misregulated genes. An exhaustive study of this misregulated response indicated that the role of NCRIP is to keep repressed a pool of phagocytosis-related genes during non-stressful conditions when there is not an interaction with macrophages. Mutant strains with an inactive NCRIP are blinded for the oxidative response, activating the genetic program against this stress when it is not present. A properly regulated genetic response to escape from macrophages is critical for the infection. In consequence, NCRIP mutants show avirulent phenotypes in murine models [11][27].
6. Summary
NCRIP is a new RNAi pathway with non-canonical machinery dependent on RdRPs and a novel RNase known as R3B2. Its main activity is to regulate the expression of target mRNAs by their degradation during specific non-stressful conditions. The most peculiar feature of NCRIP is its independence on Dicer and Argonaute proteins and their substitution with the enzyme R3B2. This novel RNase type III has a particular degrading activity because it exclusively degrades ssRNA, unlike other RNases type III that only cut dsRNA. The simplicity of the NCRIP machinery and its independence on Dicer and Argonaute proteins led to propose this non-canonical pathway as an evolutionary link between the bacterial RNA degrading pathways and the complex eukaryotic RNAi pathways [7][21][23,67]. Its machinery is similar to bacterial RNA degradation pathways, but its complex regulatory functions relate to the complex canonical RNAi pathways in eukaryotes. The regulatory functions of NCRIP include the repression of the epimutational pathway and the generation of antifungal resistant strains, the repression of the canonical RNAi in the control of transposon expression, the regulation of the fungal genetic response during phagocytosis, and the multifactorial process of virulence in Mucorales. The critical role of NCRIP in virulence and its function in regulating the antifungal resistance response make this pathway a key element in the development of mucormycosis and its treatment. Moreover, its specific conservation in Mucorales makes this non-canonical pathway an ideal target for developing new therapies against mucormycosis.
