Dysregulation of messenger RNA (mRNA) processing—in particular mRNA splicing—is a hallmark of cancer. Compared to normal cells, cancer cells frequently present aberrant mRNA splicing, which promotes cancer progression and treatment resistance. This hallmark provides opportunities for developing new targeted cancer treatments. Splicing of precursor mRNA into mature mRNA is executed by a dynamic complex of proteins and small RNAs called the spliceosome. Spliceosomes are part of the supraspliceosome, a macromolecular structure where all co-transcriptional mRNA processing activities in the cell nucleus are coordinated. Here we review the biology of the mRNA splicing machinery in the context of other mRNA processing activities in the supraspliceosome.
- alternative splicing
- splicing dysregulation
- splicing factors
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108][109][110][111][112][113][114][115][116][117][118][119][120][121][122][123][124][125][126][127][128][129][130][131][132][133][134][135][136][137][138][139][140][141][142][143][144][145][146][147][148][149][150][151][152][153][154][155][156][157][158][159][160][161][162][163][164][165][166][167][168][169][170][171][172][173][174][175][176][177][178][179][180][181][182][183][184][185][186]Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
2. Structure and Function of the (Supra) Spliceosome
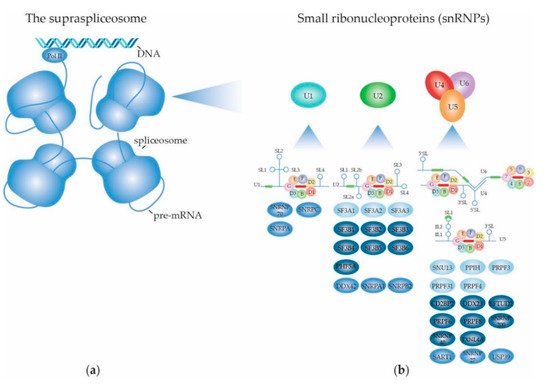
2.1. The RNA Splicing Machinery and the Splicing Reaction
2.1.1. Biogenesis of the Spliceosome: Assembly and Transport of Sm-snRNA Complexes
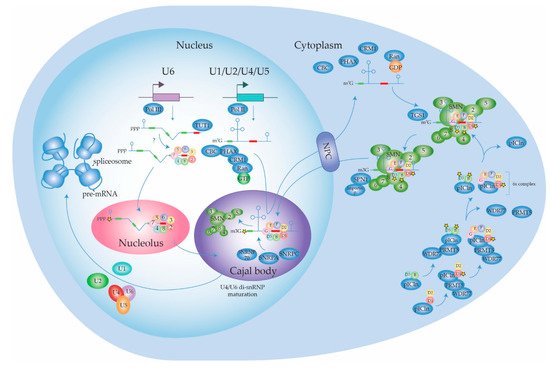
2.1.2. Biogenesis of the Spliceosome: Structure and Assembly of the U1 snRNP
2.1.3. Biogenesis of the Spliceosome: Structure and Assembly of the Other snRNPs
2.1.4. Dynamic Composition of the Spliceosome: Assembly on the pre-mRNA Substrate
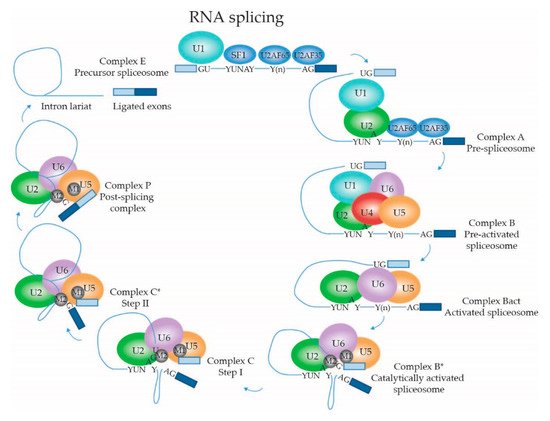
2.1.5. Dynamic Composition of the Spliceosome: Activation and Catalytic Steps
2.2. Regulation of (Alternative) mRNA Splicing
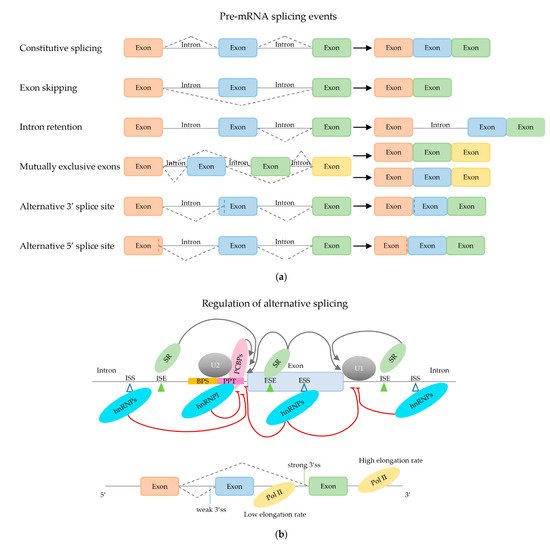
2.2.1. Trans-Acting mRNA Splicing Factors
2.2.2. Effect of Secondary mRNA Structure
2.2.3. Effect of mRNA Elongation Rate
2.2.4. Effect of Chromatin Structure
2.3. Other RNA Processing Activities in the Supraspliceosome
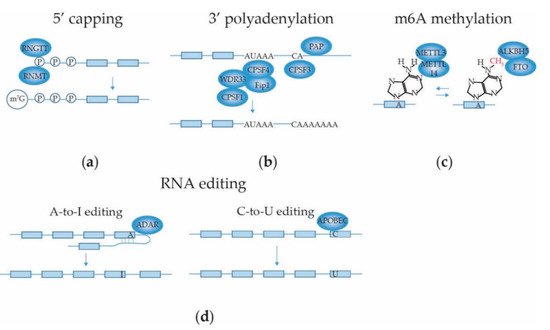
2.3.1. mRNA 5′ Capping
2.3.2. mRNA 3′ End Cleavage and Polyadenylation
2.3.3. mRNA Internal Adenosine Methylation
2.3.4. mRNA Base Editing
2.3.5. Intronic Pri-miRNAs
2.3.6. mRNA Transport
References
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175.Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175, doi:10.1073/pnas.74.8.3171.
- Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 1977, 12, 1–8.Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5’ ends of adenovirus 2 mes-senger RNA. Cell 1977, 12, 1–8, doi:10.1016/0092-8674(77)90180-5.
- Bai, R.; Yan, C.; Wan, R.; Lei, J.; Shi, Y. Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae. Cell 2017, 171, 1589–1598.e8.Bai, R.; Yan, C.; Wan, R.; Lei, J.; Shi, Y. Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae. Cell 2017, 171, 1589–1598.e8, doi:10.1016/j.cell.2017.10.038.
- Wan, R.; Yan, C.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 2016, 353, 895–904.Wan, R.; Yan, C.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 2016, 353, 895–904, doi:10.1126/science.aag2235.
- Yan, C.; Hang, J.; Wan, R.; Huang, M.; Wong, C.C.; Shi, Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015, 349, 1182–1191.Yan, C.; Hang, J.; Wan, R.; Huang, M.; Wong, C.C.; Shi, Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Sci-ence 2015, 349, 1182–1191, doi:10.1126/science.aac7629.
- Zhan, X.; Yan, C.; Zhang, X.; Lei, J.; Shi, Y. Structure of a human catalytic step I spliceosome. Science 2018, 359, 537–545.Zhan, X.; Yan, C.; Zhang, X.; Lei, J.; Shi, Y. Structure of a human catalytic step I spliceosome. Science 2018, 359, 537–545, doi:10.1126/science.aar6401.
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005, 17, 251–256.Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005, 17, 251–256, doi:10.1016/j.ceb.2005.04.006.
- Azubel, M.; Habib, N.; Sperling, R.; Sperling, J. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J. Mol. Biol. 2006, 356, 955–966.Azubel, M.; Habib, N.; Sperling, R.; Sperling, J. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J. Mol. Biol. 2006, 356, 955–966, doi:10.1016/j.jmb.2005.11.078.
- Muller, S.; Wolpensinger, B.; Angenitzki, M.; Engel, A.; Sperling, J.; Sperling, R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 1998, 283, 383–394.Muller, S.; Wolpensinger, B.; Angenitzki, M.; Engel, A.; Sperling, J.; Sperling, R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 1998, 283, 383–394, doi:10.1006/jmbi.1998.2078.
- Raitskin, O.; Angenitzki, M.; Sperling, J.; Sperling, R. Large nuclear RNP particles—The nuclear pre-mRNA processing machine. J. Struct. Biol. 2002, 140, 123–130.Raitskin, O.; Angenitzki, M.; Sperling, J.; Sperling, R. Large nuclear RNP particles—The nuclear pre-mRNA processing ma-chine. J. Struct. Biol. 2002, 140, 123–130, doi:10.1016/s1047-8477(02)00541-5.
- Raitskin, O.; Cho, D.S.; Sperling, J.; Nishikura, K.; Sperling, R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. USA 2001, 98, 6571–6576.Raitskin, O.; Cho, D.S.; Sperling, J.; Nishikura, K.; Sperling, R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. USA 2001, 98, 6571–6576, doi:10.1073/pnas.111153798.
- Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994.Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994, doi:10.1038/ncomms1998.
- Shefer, K.; Sperling, J.; Sperling, R. The Supraspliceosome—A Multi-Task Machine for Regulated Pre-mRNA Processing in the Cell Nucleus. Comput. Struct. Biotechnol. J. 2014, 11, 113–122.Shefer, K.; Sperling, J.; Sperling, R. The Supraspliceosome—A Multi-Task Machine for Regulated Pre-mRNA Processing in the Cell Nucleus. Comput. Struct. Biotechnol. J. 2014, 11, 113–122, doi:10.1016/j.csbj.2014.09.008.
- Roca, X.; Krainer, A.R.; Eperon, I.C. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013, 27, 129–144.Roca, X.; Krainer, A.R.; Eperon, I.C. Pick one, but be quick: 5’ splice sites and the problems of too many choices. Genes Dev. 2013, 27, 129–144, doi:10.1101/gad.209759.112.
- Wong, M.S.; Kinney, J.B.; Krainer, A.R. Quantitative Activity Profile and Context Dependence of All Human 5′ Splice Sites. Mol. Cell 2018, 71, 1012–1026.e3.Wong, M.S.; Kinney, J.B.; Krainer, A.R. Quantitative Activity Profile and Context Dependence of All Human 5’ Splice Sites. Mol. Cell 2018, 71, 1012–1026.e3, doi:10.1016/j.molcel.2018.07.033.
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476.Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alterna-tive isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476, doi:10.1038/nature07509.
- Tung, K.F.; Pan, C.Y.; Chen, C.H.; Lin, W.C. Top-ranked expressed gene transcripts of human protein-coding genes investigated with GTEx dataset. Sci. Rep. 2020, 10, 16245.Tung, K.F.; Pan, C.Y.; Chen, C.H.; Lin, W.C. Top-ranked expressed gene transcripts of human protein-coding genes investi-gated with GTEx dataset. Sci. Rep. 2020, 10, 16245, doi:10.1038/s41598-020-73081-5.
- Fiszbein, A.; Kornblihtt, A.R. Alternative splicing switches: Important players in cell differentiation. Bioessays 2017, 39.Fiszbein, A.; Kornblihtt, A.R. Alternative splicing switches: Important players in cell differentiation. Bioessays 2017, 39, doi:10.1002/bies.201600157.
- Maniatis, T.; Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418, 236–243.Maniatis, T.; Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418, 236–243, doi:10.1038/418236a.
- Noh, S.J.; Lee, K.; Paik, H.; Hur, C.G. TISA: Tissue-specific alternative splicing in human and mouse genes. DNA Res. 2006, 13, 229–243.Noh, S.J.; Lee, K.; Paik, H.; Hur, C.G. TISA: Tissue-specific alternative splicing in human and mouse genes. DNA Res. 2006, 13, 229–243, doi:10.1093/dnares/dsl011.
- Singh, R.K.; Cooper, T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012, 18, 472–482.Singh, R.K.; Cooper, T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012, 18, 472–482, doi:10.1016/j.molmed.2012.06.006.
- Kahles, A.; Lehmann, K.V.; Toussaint, N.C.; Huser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; The Cancer Genome Atlas Research Network; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8705 Patients. Cancer Cell 2018, 34, 211–224.e6.Kahles, A.; Lehmann, K.V.; Toussaint, N.C.; Huser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Cancer Genome Atlas Research, N.; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Pa-tients. Cancer Cell 2018, 34, 211–224.e6, doi:10.1016/j.ccell.2018.07.001.
- Sperling, R.; Koster, A.J.; Melamed-Bessudo, C.; Rubinstein, A.; Angenitzki, M.; Berkovitch-Yellin, Z.; Sperling, J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J. Mol. Biol. 1997, 267, 570–583.Sperling, R.; Koster, A.J.; Melamed-Bessudo, C.; Rubinstein, A.; Angenitzki, M.; Berkovitch-Yellin, Z.; Sperling, J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J. Mol. Biol. 1997, 267, 570–583, doi:10.1006/jmbi.1997.0898.
- Kotzer-Nevo, H.; de Lima Alves, F.; Rappsilber, J.; Sperling, J.; Sperling, R. Supraspliceosomes at defined functional states portray the pre-assembled nature of the pre-mRNA processing machine in the cell nucleus. Int. J. Mol. Sci. 2014, 15, 11637–11664.Kotzer-Nevo, H.; de Lima Alves, F.; Rappsilber, J.; Sperling, J.; Sperling, R. Supraspliceosomes at defined functional states portray the pre-assembled nature of the pre-mRNA processing machine in the cell nucleus. Int. J. Mol. Sci. 2014, 15, 11637–11664, doi:10.3390/ijms150711637.
- Cohen-Krausz, S.; Sperling, R.; Sperling, J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J. Mol. Biol. 2007, 368, 319–327.Cohen-Krausz, S.; Sperling, R.; Sperling, J. Exploring the architecture of the intact supraspliceosome using electron micros-copy. J. Mol. Biol. 2007, 368, 319–327, doi:10.1016/j.jmb.2007.01.090.
- Frankenstein, Z.; Sperling, J.; Sperling, R.; Eisenstein, M. A unique spatial arrangement of the snRNPs within the native spliceosome emerges from in silico studies. Structure 2012, 20, 1097–1106.Frankenstein, Z.; Sperling, J.; Sperling, R.; Eisenstein, M. A unique spatial arrangement of the snRNPs within the native spliceosome emerges from in silico studies. Structure 2012, 20, 1097–1106, doi:10.1016/j.str.2012.03.022.
- Sharp, P.A.; Burge, C.B. Classification of introns: U2-type or U12-type. Cell 1997, 91, 875–879.Sharp, P.A.; Burge, C.B. Classification of introns: U2-type or U12-type. Cell 1997, 91, 875–879, doi:10.1016/s0092-8674(00)80479-1.
- Fischer, U.; Englbrecht, C.; Chari, A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2011, 2, 718–731.Fischer, U.; Englbrecht, C.; Chari, A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2011, 2, 718–731, doi:10.1002/wrna.87.
- Kiss, T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004, 117, 5949–5951.Kiss, T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004, 117, 5949–5951, doi:10.1242/jcs.01487.
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121.Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121, doi:10.1038/nrm3742.
- Coady, T.H.; Lorson, C.L. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 546–564.Coady, T.H.; Lorson, C.L. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 546–564, doi:10.1002/wrna.76.
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368.Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368, doi:10.1093/nar/gkx759.
- Smith, K.P.; Hall, L.L.; Lawrence, J.B. Nuclear hubs built on RNAs and clustered organization of the genome. Curr. Opin. Cell Biol. 2020, 64, 67–76.Smith, K.P.; Hall, L.L.; Lawrence, J.B. Nuclear hubs built on RNAs and clustered organization of the genome. Curr. Opin. Cell Biol. 2020, 64, 67–76, doi:10.1016/j.ceb.2020.02.015.
- Ghaemi, Z.; Peterson, J.R.; Gruebele, M.; Luthey-Schulten, Z. An in-silico human cell model reveals the influence of spatial organization on RNA splicing. PLoS Comput. Biol. 2020, 16, e1007717.Ghaemi, Z.; Peterson, J.R.; Gruebele, M.; Luthey-Schulten, Z. An in-silico human cell model reveals the influence of spatial organization on RNA splicing. PLoS Comput. Biol. 2020, 16, e1007717, doi:10.1371/journal.pcbi.1007717.
- Veretnik, S.; Wills, C.; Youkharibache, P.; Valas, R.E.; Bourne, P.E. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput. Biol. 2009, 5, e1000315.Veretnik, S.; Wills, C.; Youkharibache, P.; Valas, R.E.; Bourne, P.E. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput. Biol. 2009, 5, e1000315, doi:10.1371/journal.pcbi.1000315.
- Lee, M.S.; Lin, Y.S.; Deng, Y.F.; Hsu, W.T.; Shen, C.C.; Cheng, Y.H.; Huang, Y.T.; Li, C. Modulation of alternative splicing by expression of small nuclear ribonucleoprotein polypeptide N. FEBS J. 2014, 281, 5194–5207.Lee, M.S.; Lin, Y.S.; Deng, Y.F.; Hsu, W.T.; Shen, C.C.; Cheng, Y.H.; Huang, Y.T.; Li, C. Modulation of alternative splicing by expression of small nuclear ribonucleoprotein polypeptide N. FEBS J. 2014, 281, 5194–5207, doi:10.1111/febs.13059.
- Hermann, H.; Fabrizio, P.; Raker, V.A.; Foulaki, K.; Hornig, H.; Brahms, H.; Luhrmann, R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995, 14, 2076–2088.
- Li, J.; Leung, A.K.; Kondo, Y.; Oubridge, C.; Nagai, K. Re-refinement of the spliceosomal U4 snRNP core-domain structure. Acta Crystallogr. D Struct. Biol. 2016, 72, 131–146.Li, J.; Leung, A.K.; Kondo, Y.; Oubridge, C.; Nagai, K. Re-refinement of the spliceosomal U4 snRNP core-domain structure. Acta Crystallogr. D Struct. Biol. 2016, 72, 131–146, doi:10.1107/S2059798315022111.
- Raker, V.A.; Hartmuth, K.; Kastner, B.; Luhrmann, R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999, 19, 6554–6565.Raker, V.A.; Hartmuth, K.; Kastner, B.; Luhrmann, R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 1999, 19, 6554–6565, doi:10.1128/mcb.19.10.6554.
- Neuenkirchen, N.; Englbrecht, C.; Ohmer, J.; Ziegenhals, T.; Chari, A.; Fischer, U. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 2015, 34, 1925–1941.Neuenkirchen, N.; Englbrecht, C.; Ohmer, J.; Ziegenhals, T.; Chari, A.; Fischer, U. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 2015, 34, 1925–1941, doi:10.15252/embj.201490350.
- Prusty, A.B.; Meduri, R.; Prusty, B.K.; Vanselow, J.; Schlosser, A.; Fischer, U. Impaired spliceosomal UsnRNP assembly leads to Sm mRNA down-regulation and Sm protein degradation. J. Cell Biol. 2017, 216, 2391–2407.Prusty, A.B.; Meduri, R.; Prusty, B.K.; Vanselow, J.; Schlosser, A.; Fischer, U. Impaired spliceosomal UsnRNP assembly leads to Sm mRNA down-regulation and Sm protein degradation. J. Cell Biol. 2017, 216, 2391–2407, doi:10.1083/jcb.201611108.
- Deng, X.; Lu, T.; Wang, L.; Gu, L.; Sun, J.; Kong, X.; Liu, C.; Cao, X. Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 2016, 113, 5447–5452.Deng, X.; Lu, T.; Wang, L.; Gu, L.; Sun, J.; Kong, X.; Liu, C.; Cao, X. Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 2016, 113, 5447–5452, doi:10.1073/pnas.1522458113.
- Gonsalvez, G.B.; Tian, L.; Ospina, J.K.; Boisvert, F.M.; Lamond, A.I.; Matera, A.G. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 2007, 178, 733–740.Gonsalvez, G.B.; Tian, L.; Ospina, J.K.; Boisvert, F.M.; Lamond, A.I.; Matera, A.G. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 2007, 178, 733–740, doi:10.1083/jcb.200702147.
- Grimm, C.; Chari, A.; Pelz, J.P.; Kuper, J.; Kisker, C.; Diederichs, K.; Stark, H.; Schindelin, H.; Fischer, U. Structural basis of assembly chaperone- mediated snRNP formation. Mol. Cell 2013, 49, 692–703.Grimm, C.; Chari, A.; Pelz, J.P.; Kuper, J.; Kisker, C.; Diederichs, K.; Stark, H.; Schindelin, H.; Fischer, U. Structural basis of assembly chaperone- mediated snRNP formation. Mol. Cell 2013, 49, 692–703, doi:10.1016/j.molcel.2012.12.009.
- Yi, H.; Mu, L.; Shen, C.; Kong, X.; Wang, Y.; Hou, Y.; Zhang, R. Negative cooperativity between Gemin2 and RNA provides insights into RNA selection and the SMN complex’s release in snRNP assembly. Nucleic Acids Res. 2020, 48, 895–911.Yi, H.; Mu, L.; Shen, C.; Kong, X.; Wang, Y.; Hou, Y.; Zhang, R. Negative cooperativity between Gemin2 and RNA provides insights into RNA selection and the SMN complex’s release in snRNP assembly. Nucleic Acids Res. 2020, 48, 895–911, doi:10.1093/nar/gkz1135.
- Ma, Y.; Dostie, J.; Dreyfuss, G.; Van Duyne, G.D. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure 2005, 13, 883–892.Ma, Y.; Dostie, J.; Dreyfuss, G.; Van Duyne, G.D. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure 2005, 13, 883–892, doi:10.1016/j.str.2005.03.014.
- Burghes, A.H.; Beattie, C.E. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009, 10, 597–609.Burghes, A.H.; Beattie, C.E. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009, 10, 597–609, doi:10.1038/nrn2670.
- Chen, T.; Zhang, B.; Ziegenhals, T.; Prusty, A.B.; Frohler, S.; Grimm, C.; Hu, Y.; Schaefke, B.; Fang, L.; Zhang, M.; et al. A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing. PLoS Genet. 2019, 15, e1008460.Chen, T.; Zhang, B.; Ziegenhals, T.; Prusty, A.B.; Frohler, S.; Grimm, C.; Hu, Y.; Schaefke, B.; Fang, L.; Zhang, M.; et al. A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing. PLoS Genet. 2019, 15, e1008460, doi:10.1371/journal.pgen.1008460.
- Jin, W.; Wang, Y.; Liu, C.P.; Yang, N.; Jin, M.; Cong, Y.; Wang, M.; Xu, R.M. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 2016, 30, 2391–2403.Jin, W.; Wang, Y.; Liu, C.P.; Yang, N.; Jin, M.; Cong, Y.; Wang, M.; Xu, R.M. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 2016, 30, 2391–2403, doi:10.1101/gad.291377.116.
- Tang, X.; Bharath, S.R.; Piao, S.; Tan, V.Q.; Bowler, M.W.; Song, H. Structural basis for specific recognition of pre-snRNA by Gemin5. Cell Res. 2016, 26, 1353–1356.Tang, X.; Bharath, S.R.; Piao, S.; Tan, V.Q.; Bowler, M.W.; Song, H. Structural basis for specific recognition of pre-snRNA by Gemin5. Cell Res. 2016, 26, 1353–1356, doi:10.1038/cr.2016.133.
- Xu, C.; Ishikawa, H.; Izumikawa, K.; Li, L.; He, H.; Nobe, Y.; Yamauchi, Y.; Shahjee, H.M.; Wu, X.H.; Yu, Y.T.; et al. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 2016, 30, 2376–2390.Xu, C.; Ishikawa, H.; Izumikawa, K.; Li, L.; He, H.; Nobe, Y.; Yamauchi, Y.; Shahjee, H.M.; Wu, X.H.; Yu, Y.T.; et al. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 2016, 30, 2376–2390, doi:10.1101/gad.288340.116.
- Yong, J.; Kasim, M.; Bachorik, J.L.; Wan, L.; Dreyfuss, G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell 2010, 38, 551–562.Yong, J.; Kasim, M.; Bachorik, J.L.; Wan, L.; Dreyfuss, G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell 2010, 38, 551–562, doi:10.1016/j.molcel.2010.03.014.
- So, B.R.; Wan, L.; Zhang, Z.; Li, P.; Babiash, E.; Duan, J.; Younis, I.; Dreyfuss, G. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 2016, 23, 225–230.So, B.R.; Wan, L.; Zhang, Z.; Li, P.; Babiash, E.; Duan, J.; Younis, I.; Dreyfuss, G. A U1 snRNP-specific assembly pathway re-veals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 2016, 23, 225–230, doi:10.1038/nsmb.3167.
- Weber, G.; Trowitzsch, S.; Kastner, B.; Luhrmann, R.; Wahl, M.C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010, 29, 4172–4184.Weber, G.; Trowitzsch, S.; Kastner, B.; Luhrmann, R.; Wahl, M.C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010, 29, 4172–4184, doi:10.1038/emboj.2010.295.
- Lund, E.; Dahlberg, J.E. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA’s. Science 1992, 255, 327–330.Lund, E.; Dahlberg, J.E. Cyclic 2’,3’-phosphates and nontemplated nucleotides at the 3’ end of spliceosomal U6 small nuclear RNA’s. Science 1992, 255, 327–330, doi:10.1126/science.1549778.
- Sontheimer, E.J.; Steitz, J.A. Three novel functional variants of human U5 small nuclear RNA. Mol. Cell Biol. 1992, 12, 734–746.Sontheimer, E.J.; Steitz, J.A. Three novel functional variants of human U5 small nuclear RNA. Mol. Cell Biol. 1992, 12, 734–746, doi:10.1128/mcb.12.2.734.
- Urlaub, H.; Raker, V.A.; Kostka, S.; Luhrmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001, 20, 187–196.Urlaub, H.; Raker, V.A.; Kostka, S.; Luhrmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceo-somal snRNP core structure. EMBO J. 2001, 20, 187–196, doi:10.1093/emboj/20.1.187.
- Schwer, B.; Kruchten, J.; Shuman, S. Structure-function analysis and genetic interactions of the SmG, SmE, and SmF subunits of the yeast Sm protein ring. RNA 2016, 22, 1320–1328.Schwer, B.; Kruchten, J.; Shuman, S. Structure-function analysis and genetic interactions of the SmG, SmE, and SmF subunits of the yeast Sm protein ring. RNA 2016, 22, 1320–1328, doi:10.1261/rna.057448.116.
- Mouaikel, J.; Narayanan, U.; Verheggen, C.; Matera, A.G.; Bertrand, E.; Tazi, J.; Bordonne, R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003, 4, 616–622.Mouaikel, J.; Narayanan, U.; Verheggen, C.; Matera, A.G.; Bertrand, E.; Tazi, J.; Bordonne, R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003, 4, 616–622, doi:10.1038/sj.embor.embor863.
- Huber, J.; Dickmanns, A.; Luhrmann, R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002, 156, 467–479.Huber, J.; Dickmanns, A.; Luhrmann, R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002, 156, 467–479, doi:10.1083/jcb.200108114.
- Roithova, A.; Klimesova, K.; Panek, J.; Will, C.L.; Luhrmann, R.; Stanek, D.; Girard, C. The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation. Nucleic Acids Res. 2018, 46, 3774–3790.Roithova, A.; Klimesova, K.; Panek, J.; Will, C.L.; Luhrmann, R.; Stanek, D.; Girard, C. The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation. Nucleic Acids Res. 2018, 46, 3774–3790, doi:10.1093/nar/gky070.
- Aoyama, T.; Yamashita, S.; Tomita, K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020, 48, 5157–5168.Aoyama, T.; Yamashita, S.; Tomita, K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nu-cleic Acids Res. 2020, 48, 5157–5168, doi:10.1093/nar/gkaa227.
- Goh, Y.T.; Koh, C.W.Q.; Sim, D.Y.; Roca, X.; Goh, W.S.S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020, 48, 9250–9261.Goh, Y.T.; Koh, C.W.Q.; Sim, D.Y.; Roca, X.; Goh, W.S.S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020, 48, 9250–9261, doi:10.1093/nar/gkaa684.
- Mauer, J.; Sindelar, M.; Despic, V.; Guez, T.; Hawley, B.R.; Vasseur, J.J.; Rentmeister, A.; Gross, S.S.; Pellizzoni, L.; Debart, F.; et al. FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019, 15, 340–347.Mauer, J.; Sindelar, M.; Despic, V.; Guez, T.; Hawley, B.R.; Vasseur, J.J.; Rentmeister, A.; Gross, S.S.; Pellizzoni, L.; Debart, F.; et al. FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019, 15, 340–347, doi:10.1038/s41589-019-0231-8.
- Pomeranz Krummel, D.A.; Oubridge, C.; Leung, A.K.; Li, J.; Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 2009, 458, 475–480.Pomeranz Krummel, D.A.; Oubridge, C.; Leung, A.K.; Li, J.; Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 2009, 458, 475–480, doi:10.1038/nature07851.
- Sun, S.; Ling, S.C.; Qiu, J.; Albuquerque, C.P.; Zhou, Y.; Tokunaga, S.; Li, H.; Qiu, H.; Bui, A.; Yeo, G.W.; et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015, 6, 6171.Sun, S.; Ling, S.C.; Qiu, J.; Albuquerque, C.P.; Zhou, Y.; Tokunaga, S.; Li, H.; Qiu, H.; Bui, A.; Yeo, G.W.; et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015, 6, 6171, doi:10.1038/ncomms7171.
- Li, X.; Liu, S.; Jiang, J.; Zhang, L.; Espinosa, S.; Hill, R.C.; Hansen, K.C.; Zhou, Z.H.; Zhao, R. CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat. Commun. 2017, 8, 1035.Li, X.; Liu, S.; Jiang, J.; Zhang, L.; Espinosa, S.; Hill, R.C.; Hansen, K.C.; Zhou, Z.H.; Zhao, R. CryoEM structure of Saccharo-myces cerevisiae U1 snRNP offers insight into alternative splicing. Nat. Commun. 2017, 8, 1035, doi:10.1038/s41467-017-01241-9.
- Nelissen, R.L.; Will, C.L.; van Venrooij, W.J.; Luhrmann, R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994, 13, 4113–4125.
- Kondo, Y.; Oubridge, C.; van Roon, A.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 2015, 4.Kondo, Y.; Oubridge, C.; van Roon, A.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleopro-tein particle, reveals the mechanism of 5’ splice site recognition. Elife 2015, 4, doi:10.7554/eLife.04986.
- Rosel-Hillgartner, T.D.; Hung, L.H.; Khrameeva, E.; Le Querrec, P.; Gelfand, M.S.; Bindereif, A. A novel intra-U1 snRNP cross-regulation mechanism: Alternative splicing switch links U1C and U1-70K expression. PLoS Genet. 2013, 9, e1003856.Rosel-Hillgartner, T.D.; Hung, L.H.; Khrameeva, E.; Le Querrec, P.; Gelfand, M.S.; Bindereif, A. A novel intra-U1 snRNP cross-regulation mechanism: Alternative splicing switch links U1C and U1-70K expression. PLoS Genet. 2013, 9, e1003856, doi:10.1371/journal.pgen.1003856.
- Zhang, Z.; Will, C.L.; Bertram, K.; Dybkov, O.; Hartmuth, K.; Agafonov, D.E.; Hofele, R.; Urlaub, H.; Kastner, B.; Luhrmann, R.; et al. Molecular architecture of the human 17S U2 snRNP. Nature 2020, 583, 310–313.Zhang, Z.; Will, C.L.; Bertram, K.; Dybkov, O.; Hartmuth, K.; Agafonov, D.E.; Hofele, R.; Urlaub, H.; Kastner, B.; Luhrmann, R.; et al. Molecular architecture of the human 17S U2 snRNP. Nature 2020, 583, 310–313, doi:10.1038/s41586-020-2344-3.
- Martelly, W.; Fellows, B.; Senior, K.; Marlowe, T.; Sharma, S. Identification of a noncanonical RNA binding domain in the U2 snRNP protein SF3A1. RNA 2019, 25, 1509–1521.Martelly, W.; Fellows, B.; Senior, K.; Marlowe, T.; Sharma, S. Identification of a noncanonical RNA binding domain in the U2 snRNP protein SF3A1. RNA 2019, 25, 1509–1521, doi:10.1261/rna.072256.119.
- Crisci, A.; Raleff, F.; Bagdiul, I.; Raabe, M.; Urlaub, H.; Rain, J.C.; Kramer, A. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015, 43, 10456–10473.Crisci, A.; Raleff, F.; Bagdiul, I.; Raabe, M.; Urlaub, H.; Rain, J.C.; Kramer, A. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015, 43, 10456–10473, doi:10.1093/nar/gkv952.
- Cretu, C.; Schmitzova, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Luhrmann, R.; Pena, V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 2016, 64, 307–319.Cretu, C.; Schmitzova, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Luhr-mann, R.; Pena, V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 2016, 64, 307–319, doi:10.1016/j.molcel.2016.08.036.
- Kfir, N.; Lev-Maor, G.; Glaich, O.; Alajem, A.; Datta, A.; Sze, S.K.; Meshorer, E.; Ast, G. SF3B1 association with chromatin determines splicing outcomes. Cell Rep. 2015, 11, 618–629.Kfir, N.; Lev-Maor, G.; Glaich, O.; Alajem, A.; Datta, A.; Sze, S.K.; Meshorer, E.; Ast, G. SF3B1 association with chromatin determines splicing outcomes. Cell Rep. 2015, 11, 618–629, doi:10.1016/j.celrep.2015.03.048.
- Sander, B.; Golas, M.M.; Makarov, E.M.; Brahms, H.; Kastner, B.; Luhrmann, R.; Stark, H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell 2006, 24, 267–278.Sander, B.; Golas, M.M.; Makarov, E.M.; Brahms, H.; Kastner, B.; Luhrmann, R.; Stark, H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell 2006, 24, 267–278, doi:10.1016/j.molcel.2006.08.021.
- Haselbach, D.; Komarov, I.; Agafonov, D.E.; Hartmuth, K.; Graf, B.; Dybkov, O.; Urlaub, H.; Kastner, B.; Luhrmann, R.; Stark, H. Structure and Conformational Dynamics of the Human Spliceosomal B(act) Complex. Cell 2018, 172, 454–464.e11.Haselbach, D.; Komarov, I.; Agafonov, D.E.; Hartmuth, K.; Graf, B.; Dybkov, O.; Urlaub, H.; Kastner, B.; Luhrmann, R.; Stark, H. Structure and Conformational Dynamics of the Human Spliceosomal B(act) Complex. Cell 2018, 172, 454–464.e11, doi:10.1016/j.cell.2018.01.010.
- Agafonov, D.E.; Kastner, B.; Dybkov, O.; Hofele, R.V.; Liu, W.T.; Urlaub, H.; Luhrmann, R.; Stark, H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016, 351, 1416–1420.Agafonov, D.E.; Kastner, B.; Dybkov, O.; Hofele, R.V.; Liu, W.T.; Urlaub, H.; Luhrmann, R.; Stark, H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016, 351, 1416–1420, doi:10.1126/science.aad2085.
- Hardin, J.W.; Warnasooriya, C.; Kondo, Y.; Nagai, K.; Rueda, D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 2015, 43, 10963–10974.Hardin, J.W.; Warnasooriya, C.; Kondo, Y.; Nagai, K.; Rueda, D. Assembly and dynamics of the U4/U6 di-snRNP by sin-gle-molecule FRET. Nucleic Acids Res. 2015, 43, 10963–10974, doi:10.1093/nar/gkv1011.
- Cloutier, P.; Poitras, C.; Durand, M.; Hekmat, O.; Fiola-Masson, E.; Bouchard, A.; Faubert, D.; Chabot, B.; Coulombe, B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017, 8, 15615.Cloutier, P.; Poitras, C.; Durand, M.; Hekmat, O.; Fiola-Masson, E.; Bouchard, A.; Faubert, D.; Chabot, B.; Coulombe, B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017, 8, 15615, doi:10.1038/ncomms15615.
- Malinova, A.; Cvackova, Z.; Mateju, D.; Horejsi, Z.; Abeza, C.; Vandermoere, F.; Bertrand, E.; Stanek, D.; Verheggen, C. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017, 216, 1579–1596.Malinova, A.; Cvackova, Z.; Mateju, D.; Horejsi, Z.; Abeza, C.; Vandermoere, F.; Bertrand, E.; Stanek, D.; Verheggen, C. As-sembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017, 216, 1579–1596, doi:10.1083/jcb.201701165.
- Yamashita, S.; Takagi, Y.; Nagaike, T.; Tomita, K. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 2017, 8, 15788.Yamashita, S.; Takagi, Y.; Nagaike, T.; Tomita, K. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 2017, 8, 15788, doi:10.1038/ncomms15788.
- Achsel, T.; Brahms, H.; Kastner, B.; Bachi, A.; Wilm, M.; Luhrmann, R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999, 18, 5789–5802.Achsel, T.; Brahms, H.; Kastner, B.; Bachi, A.; Wilm, M.; Luhrmann, R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3’-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999, 18, 5789–5802, doi:10.1093/emboj/18.20.5789.
- Zhou, L.; Hang, J.; Zhou, Y.; Wan, R.; Lu, G.; Yin, P.; Yan, C.; Shi, Y. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 2014, 506, 116–120.Zhou, L.; Hang, J.; Zhou, Y.; Wan, R.; Lu, G.; Yin, P.; Yan, C.; Shi, Y. Crystal structures of the Lsm complex bound to the 3’ end sequence of U6 small nuclear RNA. Nature 2014, 506, 116–120, doi:10.1038/nature12803.
- Montemayor, E.J.; Didychuk, A.L.; Yake, A.D.; Sidhu, G.K.; Brow, D.A.; Butcher, S.E. Architecture of the U6 snRNP reveals specific recognition of 3′-end processed U6 snRNA. Nat. Commun. 2018, 9, 1749.Montemayor, E.J.; Didychuk, A.L.; Yake, A.D.; Sidhu, G.K.; Brow, D.A.; Butcher, S.E. Architecture of the U6 snRNP reveals specific recognition of 3’-end processed U6 snRNA. Nat. Commun. 2018, 9, 1749, doi:10.1038/s41467-018-04145-4.
- Mund, M.; Neu, A.; Ullmann, J.; Neu, U.; Sprangers, R. Structure of the LSm657 complex: An assembly intermediate of the LSm1-7 and LSm2-8 rings. J. Mol. Biol. 2011, 414, 165–176.Mund, M.; Neu, A.; Ullmann, J.; Neu, U.; Sprangers, R. Structure of the LSm657 complex: An assembly intermediate of the LSm1-7 and LSm2-8 rings. J. Mol. Biol. 2011, 414, 165–176, doi:10.1016/j.jmb.2011.09.051.
- Brahms, H.; Meheus, L.; de Brabandere, V.; Fischer, U.; Luhrmann, R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 2001, 7, 1531–1542.Brahms, H.; Meheus, L.; de Brabandere, V.; Fischer, U.; Luhrmann, R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 2001, 7, 1531–1542, doi:10.1017/s135583820101442x.
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670.Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670, doi:10.1038/nrm.2017.86.
- Wan, R.; Bai, R.; Shi, Y. Molecular choreography of pre-mRNA splicing by the spliceosome. Curr. Opin. Struct. Biol. 2019, 59, 124–133.Wan, R.; Bai, R.; Shi, Y. Molecular choreography of pre-mRNA splicing by the spliceosome. Curr. Opin. Struct. Biol. 2019, 59, 124–133, doi:10.1016/j.sbi.2019.07.010.
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect Biol. 2011, 3.Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect Biol. 2011, 3, doi:10.1101/cshperspect.a003707.
- Robberson, B.L.; Cote, G.J.; Berget, S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell Biol. 1990, 10, 84–94.Robberson, B.L.; Cote, G.J.; Berget, S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell Biol. 1990, 10, 84–94, doi:10.1128/mcb.10.1.84.
- Schwer, B.; Shuman, S. Structure-function analysis of the Yhc1 subunit of yeast U1 snRNP and genetic interactions of Yhc1 with Mud2, Nam8, Mud1, Tgs1, U1 snRNA, SmD3 and Prp28. Nucleic Acids Res. 2014, 42, 4697–4711.Schwer, B.; Shuman, S. Structure-function analysis of the Yhc1 subunit of yeast U1 snRNP and genetic interactions of Yhc1 with Mud2, Nam8, Mud1, Tgs1, U1 snRNA, SmD3 and Prp28. Nucleic Acids Res. 2014, 42, 4697–4711, doi:10.1093/nar/gku097.
- Das, R.; Yu, J.; Zhang, Z.; Gygi, M.P.; Krainer, A.R.; Gygi, S.P.; Reed, R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 2007, 26, 867–881.Das, R.; Yu, J.; Zhang, Z.; Gygi, M.P.; Krainer, A.R.; Gygi, S.P.; Reed, R. SR proteins function in coupling RNAP II transcrip-tion to pre-mRNA splicing. Mol. Cell 2007, 26, 867–881, doi:10.1016/j.molcel.2007.05.036.
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380.Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380, doi:10.1038/s41586-019-1523-6.
- Lockhart, S.R.; Rymond, B.C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 1994, 14, 3623–3633.Lockhart, S.R.; Rymond, B.C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 1994, 14, 3623–3633, doi:10.1128/mcb.14.6.3623.
- Briese, M.; Haberman, N.; Sibley, C.R.; Faraway, R.; Elser, A.S.; Chakrabarti, A.M.; Wang, Z.; Konig, J.; Perera, D.; Wickramasinghe, V.O.; et al. A systems view of spliceosomal assembly and branchpoints with iCLIP. Nat. Struct. Mol. Biol. 2019, 26, 930–940.Briese, M.; Haberman, N.; Sibley, C.R.; Faraway, R.; Elser, A.S.; Chakrabarti, A.M.; Wang, Z.; Konig, J.; Perera, D.; Wick-ramasinghe, V.O.; et al. A systems view of spliceosomal assembly and branchpoints with iCLIP. Nat. Struct. Mol. Biol. 2019, 26, 930–940, doi:10.1038/s41594-019-0300-4.
- Zarnack, K.; Konig, J.; Tajnik, M.; Martincorena, I.; Eustermann, S.; Stevant, I.; Reyes, A.; Anders, S.; Luscombe, N.M.; Ule, J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013, 152, 453–466.Zarnack, K.; Konig, J.; Tajnik, M.; Martincorena, I.; Eustermann, S.; Stevant, I.; Reyes, A.; Anders, S.; Luscombe, N.M.; Ule, J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013, 152, 453–466, doi:10.1016/j.cell.2012.12.023.
- Plaschka, C.; Lin, P.C.; Charenton, C.; Nagai, K. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 2018, 559, 419–422.Plaschka, C.; Lin, P.C.; Charenton, C.; Nagai, K. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 2018, 559, 419–422, doi:10.1038/s41586-018-0323-8.
- Sharma, S.; Kohlstaedt, L.A.; Damianov, A.; Rio, D.C.; Black, D.L. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008, 15, 183–191.Sharma, S.; Kohlstaedt, L.A.; Damianov, A.; Rio, D.C.; Black, D.L. Polypyrimidine tract binding protein controls the transi-tion from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008, 15, 183–191, doi:10.1038/nsmb.1375.
- Charenton, C.; Wilkinson, M.E.; Nagai, K. Mechanism of 5′ splice site transfer for human spliceosome activation. Science 2019, 364, 362–367.Charenton, C.; Wilkinson, M.E.; Nagai, K. Mechanism of 5’ splice site transfer for human spliceosome activation. Science 2019, 364, 362–367, doi:10.1126/science.aax3289.
- Huang, Y.H.; Chung, C.S.; Kao, D.I.; Kao, T.C.; Cheng, S.C. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol. Cell. Biol. 2014, 34, 210–220.Huang, Y.H.; Chung, C.S.; Kao, D.I.; Kao, T.C.; Cheng, S.C. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol. Cell. Biol. 2014, 34, 210–220, doi:10.1128/MCB.00837-13.
- Boesler, C.; Rigo, N.; Anokhina, M.M.; Tauchert, M.J.; Agafonov, D.E.; Kastner, B.; Urlaub, H.; Ficner, R.; Will, C.L.; Luhrmann, R. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat. Commun. 2016, 7, 11997.Boesler, C.; Rigo, N.; Anokhina, M.M.; Tauchert, M.J.; Agafonov, D.E.; Kastner, B.; Urlaub, H.; Ficner, R.; Will, C.L.; Luhr-mann, R. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activi-ty. Nat. Commun. 2016, 7, 11997, doi:10.1038/ncomms11997.
- de Almeida, R.A.; O’Keefe, R.T. The NineTeen Complex (NTC) and NTC-associated proteins as targets for spliceosomal ATPase action during pre-mRNA splicing. RNA Biol. 2015, 12, 109–114.de Almeida, R.A.; O’Keefe, R.T. The NineTeen Complex (NTC) and NTC-associated proteins as targets for spliceosomal ATPase action during pre-mRNA splicing. RNA Biol. 2015, 12, 109–114, doi:10.1080/15476286.2015.1008926.
- Hogg, R.; McGrail, J.C.; O’Keefe, R.T. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 2010, 38, 1110–1115.Hogg, R.; McGrail, J.C.; O’Keefe, R.T. The function of the NineTeen Complex (NTC) in regulating spliceosome confor-mations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 2010, 38, 1110–1115, doi:10.1042/BST0381110.
- Nguyen, T.H.; Galej, W.P.; Bai, X.C.; Savva, C.G.; Newman, A.J.; Scheres, S.H.; Nagai, K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015, 523, 47–52.Nguyen, T.H.; Galej, W.P.; Bai, X.C.; Savva, C.G.; Newman, A.J.; Scheres, S.H.; Nagai, K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015, 523, 47–52, doi:10.1038/nature14548.
- Zahler, A.M.; Rogel, L.E.; Glover, M.L.; Yitiz, S.; Ragle, J.M.; Katzman, S. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5′ splice site positioning in the spliceosome. RNA 2018, 24, 1314–1325.Zahler, A.M.; Rogel, L.E.; Glover, M.L.; Yitiz, S.; Ragle, J.M.; Katzman, S. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5’ splice site positioning in the spliceosome. RNA 2018, 24, 1314–1325, doi:10.1261/rna.066878.118.
- Townsend, C.; Leelaram, M.N.; Agafonov, D.E.; Dybkov, O.; Will, C.L.; Bertram, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhrmann, R. Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation. Science 2020, 370.Townsend, C.; Leelaram, M.N.; Agafonov, D.E.; Dybkov, O.; Will, C.L.; Bertram, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhr-mann, R. Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation. Science 2020, 370, doi:10.1126/science.abc3753.
- Hang, J.; Wan, R.; Yan, C.; Shi, Y. Structural basis of pre-mRNA splicing. Science 2015, 349, 1191–1198.Hang, J.; Wan, R.; Yan, C.; Shi, Y. Structural basis of pre-mRNA splicing. Science 2015, 349, 1191–1198, doi:10.1126/science.aac8159.
- Rauhut, R.; Fabrizio, P.; Dybkov, O.; Hartmuth, K.; Pena, V.; Chari, A.; Kumar, V.; Lee, C.T.; Urlaub, H.; Kastner, B.; et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 2016, 353, 1399–1405.Rauhut, R.; Fabrizio, P.; Dybkov, O.; Hartmuth, K.; Pena, V.; Chari, A.; Kumar, V.; Lee, C.T.; Urlaub, H.; Kastner, B.; et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 2016, 353, 1399–1405, doi:10.1126/science.aag1906.
- Sun, C.; Rigo, N.; Fabrizio, P.; Kastner, B.; Luhrmann, R. A protein map of the yeast activated spliceosome as obtained by electron microscopy. RNA 2016, 22, 1427–1440.Sun, C.; Rigo, N.; Fabrizio, P.; Kastner, B.; Luhrmann, R. A protein map of the yeast activated spliceosome as obtained by electron microscopy. RNA 2016, 22, 1427–1440, doi:10.1261/rna.057778.116.
- Bao, P.; Boon, K.L.; Will, C.L.; Hartmuth, K.; Luhrmann, R. Multiple RNA-RNA tertiary interactions are dispensable for formation of a functional U2/U6 RNA catalytic core in the spliceosome. Nucleic Acids Res. 2018, 46, 12126–12138.Bao, P.; Boon, K.L.; Will, C.L.; Hartmuth, K.; Luhrmann, R. Multiple RNA-RNA tertiary interactions are dispensable for formation of a functional U2/U6 RNA catalytic core in the spliceosome. Nucleic Acids Res. 2018, 46, 12126–12138, doi:10.1093/nar/gky966.
- Galej, W.P.; Wilkinson, M.E.; Fica, S.M.; Oubridge, C.; Newman, A.J.; Nagai, K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016, 537, 197–201.Galej, W.P.; Wilkinson, M.E.; Fica, S.M.; Oubridge, C.; Newman, A.J.; Nagai, K. Cryo-EM structure of the spliceosome im-mediately after branching. Nature 2016, 537, 197–201, doi:10.1038/nature19316.
- Zhang, X.; Yan, C.; Zhan, X.; Li, L.; Lei, J.; Shi, Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018, 28, 307–322.Zhang, X.; Yan, C.; Zhan, X.; Li, L.; Lei, J.; Shi, Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018, 28, 307–322, doi:10.1038/cr.2018.14.
- Yan, C.; Wan, R.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast step II catalytically activated spliceosome. Science 2017, 355, 149–155.Yan, C.; Wan, R.; Bai, R.; Huang, G.; Shi, Y. Structure of a yeast step II catalytically activated spliceosome. Science 2017, 355, 149–155, doi:10.1126/science.aak9979.
- Wan, R.; Bai, R.; Yan, C.; Lei, J.; Shi, Y. Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching. Cell 2019, 177, 339–351.e13.Wan, R.; Bai, R.; Yan, C.; Lei, J.; Shi, Y. Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching. Cell 2019, 177, 339–351.e13, doi:10.1016/j.cell.2019.02.006.
- Chung, C.S.; Tseng, C.K.; Lai, Y.H.; Wang, H.F.; Newman, A.J.; Cheng, S.C. Dynamic protein-RNA interactions in mediating splicing catalysis. Nucleic Acids Res. 2019, 47, 899–910.Chung, C.S.; Tseng, C.K.; Lai, Y.H.; Wang, H.F.; Newman, A.J.; Cheng, S.C. Dynamic protein-RNA interactions in mediating splicing catalysis. Nucleic Acids Res. 2019, 47, 899–910, doi:10.1093/nar/gky1089.
- Gehring, N.H.; Lamprinaki, S.; Hentze, M.W.; Kulozik, A.E. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009, 7, e1000120.Gehring, N.H.; Lamprinaki, S.; Hentze, M.W.; Kulozik, A.E. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009, 7, e1000120, doi:10.1371/journal.pbio.1000120.
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866.Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866, doi:10.3390/biom10060866.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108, doi:10.1038/nature11233.
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451.Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451, doi:10.1038/nrm.2017.27.
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165.Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165, doi:10.1038/nrm3525.
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345.Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345, doi:10.1016/j.molcel.2019.09.017.
- Shen, M.; Mattox, W. Activation and repression functions of an SR splicing regulator depend on exonic versus intronic-binding position. Nucleic Acids Res. 2012, 40, 428–437.Shen, M.; Mattox, W. Activation and repression functions of an SR splicing regulator depend on exonic versus intron-ic-binding position. Nucleic Acids Res. 2012, 40, 428–437, doi:10.1093/nar/gkr713.
- Bradley, T.; Cook, M.E.; Blanchette, M. SR proteins control a complex network of RNA-processing events. RNA 2015, 21, 75–92.Bradley, T.; Cook, M.E.; Blanchette, M. SR proteins control a complex network of RNA-processing events. RNA 2015, 21, 75–92, doi:10.1261/rna.043893.113.
- Pandit, S.; Zhou, Y.; Shiue, L.; Coutinho-Mansfield, G.; Li, H.; Qiu, J.; Huang, J.; Yeo, G.W.; Ares, M., Jr.; Fu, X.D. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 2013, 50, 223–235.Pandit, S.; Zhou, Y.; Shiue, L.; Coutinho-Mansfield, G.; Li, H.; Qiu, J.; Huang, J.; Yeo, G.W.; Ares, M., Jr.; Fu, X.D. Ge-nome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 2013, 50, 223–235, doi:10.1016/j.molcel.2013.03.001.
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236.Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236, doi:10.1186/gb-2011-12-12-236.
- Payer, L.M.; Steranka, J.P.; Ardeljan, D.; Walker, J.; Fitzgerald, K.C.; Calabresi, P.A.; Cooper, T.A.; Burns, K.H. Alu insertion variants alter mRNA splicing. Nucleic Acids Res. 2019, 47, 421–431.Payer, L.M.; Steranka, J.P.; Ardeljan, D.; Walker, J.; Fitzgerald, K.C.; Calabresi, P.A.; Cooper, T.A.; Burns, K.H. Alu insertion variants alter mRNA splicing. Nucleic Acids Res. 2019, 47, 421–431, doi:10.1093/nar/gky1086.
- Loh, T.J.; Cho, S.; Moon, H.; Jang, H.N.; Williams, D.R.; Jung, D.W.; Kim, I.C.; Ghigna, C.; Biamonti, G.; Zheng, X.; et al. hnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochim. Biophys. Acta 2015, 1849, 743–750.Loh, T.J.; Cho, S.; Moon, H.; Jang, H.N.; Williams, D.R.; Jung, D.W.; Kim, I.C.; Ghigna, C.; Biamonti, G.; Zheng, X.; et al. hnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochim. Biophys. Acta 2015, 1849, 743–750, doi:10.1016/j.bbagrm.2015.01.004.
- Ji, X.; Park, J.W.; Bahrami-Samani, E.; Lin, L.; Duncan-Lewis, C.; Pherribo, G.; Xing, Y.; Liebhaber, S.A. alphaCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 2016, 44, 2283–2297.Ji, X.; Park, J.W.; Bahrami-Samani, E.; Lin, L.; Duncan-Lewis, C.; Pherribo, G.; Xing, Y.; Liebhaber, S.A. alphaCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcrip-tome. Nucleic Acids Res. 2016, 44, 2283–2297, doi:10.1093/nar/gkw088.
- Xiao, X.; Wang, Z.; Jang, M.; Nutiu, R.; Wang, E.T.; Burge, C.B. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat. Struct Mol. Biol. 2009, 16, 1094–1100.Xiao, X.; Wang, Z.; Jang, M.; Nutiu, R.; Wang, E.T.; Burge, C.B. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat. Struct Mol. Biol. 2009, 16, 1094–1100, doi:10.1038/nsmb.1661.
- Chen, C.D.; Kobayashi, R.; Helfman, D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999, 13, 593–606.Chen, C.D.; Kobayashi, R.; Helfman, D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999, 13, 593–606, doi:10.1101/gad.13.5.593.
- Jamison, S.F.; Pasman, Z.; Wang, J.; Will, C.; Luhrmann, R.; Manley, J.L.; Garcia-Blanco, M.A. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: Characterization of required elements. Nucleic Acids Res. 1995, 23, 3260–3267.Jamison, S.F.; Pasman, Z.; Wang, J.; Will, C.; Luhrmann, R.; Manley, J.L.; Garcia-Blanco, M.A. U1 snRNP-ASF/SF2 interaction and 5’ splice site recognition: Characterization of required elements. Nucleic Acids Res. 1995, 23, 3260–3267, doi:10.1093/nar/23.16.3260.
- Cho, S.; Hoang, A.; Sinha, R.; Zhong, X.Y.; Fu, X.D.; Krainer, A.R.; Ghosh, G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 8233–8238.Cho, S.; Hoang, A.; Sinha, R.; Zhong, X.Y.; Fu, X.D.; Krainer, A.R.; Ghosh, G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 8233–8238, doi:10.1073/pnas.1017700108.
- Jobbins, A.M.; Reichenbach, L.F.; Lucas, C.M.; Hudson, A.J.; Burley, G.A.; Eperon, I.C. The mechanisms of a mammalian splicing enhancer. Nucleic Acids Res. 2018, 46, 2145–2158.Jobbins, A.M.; Reichenbach, L.F.; Lucas, C.M.; Hudson, A.J.; Burley, G.A.; Eperon, I.C. The mechanisms of a mammalian splicing enhancer. Nucleic Acids Res. 2018, 46, 2145–2158, doi:10.1093/nar/gky056.
- Aubol, B.E.; Wu, G.; Keshwani, M.M.; Movassat, M.; Fattet, L.; Hertel, K.J.; Fu, X.D.; Adams, J.A. Release of SR Proteins from CLK1 by SRPK1: A Symbiotic Kinase System for Phosphorylation Control of Pre-mRNA Splicing. Mol. Cell 2016, 63, 218–228.Aubol, B.E.; Wu, G.; Keshwani, M.M.; Movassat, M.; Fattet, L.; Hertel, K.J.; Fu, X.D.; Adams, J.A. Release of SR Proteins from CLK1 by SRPK1: A Symbiotic Kinase System for Phosphorylation Control of Pre-mRNA Splicing. Mol. Cell 2016, 63, 218–228, doi:10.1016/j.molcel.2016.05.034.
- Keiper, S.; Papasaikas, P.; Will, C.L.; Valcarcel, J.; Girard, C.; Luhrmann, R. Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns. Nat. Commun. 2019, 10, 3639.Keiper, S.; Papasaikas, P.; Will, C.L.; Valcarcel, J.; Girard, C.; Luhrmann, R. Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns. Nat. Commun. 2019, 10, 3639, doi:10.1038/s41467-019-11293-8.
- Papasaikas, P.; Tejedor, J.R.; Vigevani, L.; Valcarcel, J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell 2015, 57, 7–22.Papasaikas, P.; Tejedor, J.R.; Vigevani, L.; Valcarcel, J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell 2015, 57, 7–22, doi:10.1016/j.molcel.2014.10.030.
- Subramania, S.; Gagne, L.M.; Campagne, S.; Fort, V.; O’Sullivan, J.; Mocaer, K.; Feldmuller, M.; Masson, J.Y.; Allain, F.H.T.; Hussein, S.M.; et al. SAM68 interaction with U1A modulates U1 snRNP recruitment and regulates mTor pre-mRNA splicing. Nucleic Acids Res. 2019, 47, 4181–4197.Subramania, S.; Gagne, L.M.; Campagne, S.; Fort, V.; O’Sullivan, J.; Mocaer, K.; Feldmuller, M.; Masson, J.Y.; Allain, F.H.T.; Hussein, S.M.; et al. SAM68 interaction with U1A modulates U1 snRNP recruitment and regulates mTor pre-mRNA splicing. Nucleic Acids Res. 2019, 47, 4181–4197, doi:10.1093/nar/gkz099.
- Adams, B.M.; Coates, M.N.; Jackson, S.R.; Jurica, M.S.; Davis, T.L. Nuclear cyclophilins affect spliceosome assembly and function in vitro. Biochem. J. 2015, 469, 223–233.Adams, B.M.; Coates, M.N.; Jackson, S.R.; Jurica, M.S.; Davis, T.L. Nuclear cyclophilins affect spliceosome assembly and function in vitro. Biochem. J. 2015, 469, 223–233, doi:10.1042/BJ20150396.
- Bartys, N.; Kierzek, R.; Lisowiec-Wachnicka, J. The regulation properties of RNA secondary structure in alternative splicing. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194401.Bartys, N.; Kierzek, R.; Lisowiec-Wachnicka, J. The regulation properties of RNA secondary structure in alternative splicing. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194401, doi:10.1016/j.bbagrm.2019.07.002.
- Warf, M.B.; Diegel, J.V.; von Hippel, P.H.; Berglund, J.A. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 9203–9208.Warf, M.B.; Diegel, J.V.; von Hippel, P.H.; Berglund, J.A. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 9203–9208, doi:10.1073/pnas.0900342106.
- von Hacht, A.; Seifert, O.; Menger, M.; Schutze, T.; Arora, A.; Konthur, Z.; Neubauer, P.; Wagner, A.; Weise, C.; Kurreck, J. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014, 42, 6630–6644.von Hacht, A.; Seifert, O.; Menger, M.; Schutze, T.; Arora, A.; Konthur, Z.; Neubauer, P.; Wagner, A.; Weise, C.; Kurreck, J. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014, 42, 6630–6644, doi:10.1093/nar/gku290.
- Conlon, E.G.; Lu, L.; Sharma, A.; Yamazaki, T.; Tang, T.; Shneider, N.A.; Manley, J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 2016, 5.Conlon, E.G.; Lu, L.; Sharma, A.; Yamazaki, T.; Tang, T.; Shneider, N.A.; Manley, J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 2016, 5, doi:10.7554/eLife.17820.
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309.Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309, doi:10.1101/gad.305862.117.
- Baraniak, A.P.; Lasda, E.L.; Wagner, E.J.; Garcia-Blanco, M.A. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003, 23, 9327–9337.Baraniak, A.P.; Lasda, E.L.; Wagner, E.J.; Garcia-Blanco, M.A. A stem structure in fibroblast growth factor receptor 2 tran-scripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003, 23, 9327–9337, doi:10.1128/mcb.23.24.9327-9337.2003.
- Munding, E.M.; Shiue, L.; Katzman, S.; Donohue, J.P.; Ares, M., Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell 2013, 51, 338–348.Munding, E.M.; Shiue, L.; Katzman, S.; Donohue, J.P.; Ares, M., Jr. Competition between pre-mRNAs for the splicing ma-chinery drives global regulation of splicing. Mol. Cell 2013, 51, 338–348, doi:10.1016/j.molcel.2013.06.012.
- Ding, F.; Elowitz, M.B. Constitutive splicing and economies of scale in gene expression. Nat. Struct. Mol. Biol. 2019, 26, 424–432.Ding, F.; Elowitz, M.B. Constitutive splicing and economies of scale in gene expression. Nat. Struct. Mol. Biol. 2019, 26, 424–432, doi:10.1038/s41594-019-0226-x.
- Alexander, R.D.; Innocente, S.A.; Barrass, J.D.; Beggs, J.D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell 2010, 40, 582–593.Alexander, R.D.; Innocente, S.A.; Barrass, J.D.; Beggs, J.D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell 2010, 40, 582–593, doi:10.1016/j.molcel.2010.11.005.
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478.Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478, doi:10.1038/s41580-018-0010-5.
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem 2015, 84, 165–198.Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem 2015, 84, 165–198, doi:10.1146/annurev-biochem-060614-034242.
- Fong, N.; Kim, H.; Zhou, Y.; Ji, X.; Qiu, J.; Saldi, T.; Diener, K.; Jones, K.; Fu, X.D.; Bentley, D.L. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014, 28, 2663–2676.Fong, N.; Kim, H.; Zhou, Y.; Ji, X.; Qiu, J.; Saldi, T.; Diener, K.; Jones, K.; Fu, X.D.; Bentley, D.L. Pre-mRNA splicing is facili-tated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014, 28, 2663–2676, doi:10.1101/gad.252106.114.
- Baluapuri, A.; Hofstetter, J.; Dudvarski Stankovic, N.; Endres, T.; Bhandare, P.; Vos, S.M.; Adhikari, B.; Schwarz, J.D.; Narain, A.; Vogt, M.; et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol. Cell 2019, 74, 674–687.e11.Baluapuri, A.; Hofstetter, J.; Dudvarski Stankovic, N.; Endres, T.; Bhandare, P.; Vos, S.M.; Adhikari, B.; Schwarz, J.D.; Narain, A.; Vogt, M.; et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol. Cell 2019, 74, 674–687.e11, doi:10.1016/j.molcel.2019.02.031.
- Lin, C.Y.; Loven, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67.Lin, C.Y.; Loven, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67, doi:10.1016/j.cell.2012.08.026.
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26.Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26, doi:10.1016/j.cell.2010.11.056.
- Narayanan, S.P.; Singh, S.; Shukla, S. A saga of cancer epigenetics: Linking epigenetics to alternative splicing. Biochem. J. 2017, 474, 885–896.Narayanan, S.P.; Singh, S.; Shukla, S. A saga of cancer epigenetics: Linking epigenetics to alternative splicing. Biochem. J. 2017, 474, 885–896, doi:10.1042/BCJ20161047.
- Schwartz, S.; Meshorer, E.; Ast, G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009, 16, 990–995.Schwartz, S.; Meshorer, E.; Ast, G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009, 16, 990–995, doi:10.1038/nsmb.1659.
- Spies, N.; Nielsen, C.B.; Padgett, R.A.; Burge, C.B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 2009, 36, 245–254.Spies, N.; Nielsen, C.B.; Padgett, R.A.; Burge, C.B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 2009, 36, 245–254, doi:10.1016/j.molcel.2009.10.008.
- Sims, R.J., 3rd; Millhouse, S.; Chen, C.F.; Lewis, B.A.; Erdjument-Bromage, H.; Tempst, P.; Manley, J.L.; Reinberg, D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 2007, 28, 665–676.Sims, R.J., 3rd; Millhouse, S.; Chen, C.F.; Lewis, B.A.; Erdjument-Bromage, H.; Tempst, P.; Manley, J.L.; Reinberg, D. Recog-nition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 2007, 28, 665–676, doi:10.1016/j.molcel.2007.11.010.
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000.Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by his-tone modifications. Science 2010, 327, 996–1000, doi:10.1126/science.1184208.
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526.Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526, doi:10.1093/nar/gkw551.
- Kumar, A.; Clerici, M.; Muckenfuss, L.M.; Passmore, L.A.; Jinek, M. Mechanistic insights into mRNA 3′-end processing. Curr. Opin. Struct. Biol. 2019, 59, 143–150.Kumar, A.; Clerici, M.; Muckenfuss, L.M.; Passmore, L.A.; Jinek, M. Mechanistic insights into mRNA 3’-end processing. Curr. Opin. Struct. Biol. 2019, 59, 143–150, doi:10.1016/j.sbi.2019.08.001.
- Chen, Y.I.; Moore, R.E.; Ge, H.Y.; Young, M.K.; Lee, T.D.; Stevens, S.W. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007, 35, 3928–3944.Chen, Y.I.; Moore, R.E.; Ge, H.Y.; Young, M.K.; Lee, T.D.; Stevens, S.W. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007, 35, 3928–3944, doi:10.1093/nar/gkm347.
- Berg, M.G.; Singh, L.N.; Younis, I.; Liu, Q.; Pinto, A.M.; Kaida, D.; Zhang, Z.; Cho, S.; Sherrill-Mix, S.; Wan, L.; et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 2012, 150, 53–64.Berg, M.G.; Singh, L.N.; Younis, I.; Liu, Q.; Pinto, A.M.; Kaida, D.; Zhang, Z.; Cho, S.; Sherrill-Mix, S.; Wan, L.; et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 2012, 150, 53–64, doi:10.1016/j.cell.2012.05.029.
- Kaida, D.; Berg, M.G.; Younis, I.; Kasim, M.; Singh, L.N.; Wan, L.; Dreyfuss, G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010, 468, 664–668.Kaida, D.; Berg, M.G.; Younis, I.; Kasim, M.; Singh, L.N.; Wan, L.; Dreyfuss, G. U1 snRNP protects pre-mRNAs from prema-ture cleavage and polyadenylation. Nature 2010, 468, 664–668, doi:10.1038/nature09479.
- He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977.He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977, doi:10.15252/embj.2020105977.
- Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 2020, 19, 88.Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 2020, 19, 88, doi:10.1186/s12943-020-01204-7.
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564.Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564, doi:10.1038/nature14234.
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063.Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate bind-ing of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063, doi:10.1093/nar/gkx141.
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419.Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419, doi:10.1038/cr.2014.151.
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29.Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29, doi:10.1016/j.molcel.2012.10.015.
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519.Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519, doi:10.1016/j.molcel.2016.01.012.
- Samuel, C.E. RNA Editing. In Reference Module in Biomedical Sciences; Caplan, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–6.Samuel, C.E. RNA Editing. In Reference Module in Biomedical Sciences; Caplan, M., edEd.; Elsevier: Amsterdam, The Nether-lands, 2019; pp 1-6, doi:10.1016/B978-0-12-801238-3.11364-9.
- Phelps, K.J.; Tran, K.; Eifler, T.; Erickson, A.I.; Fisher, A.J.; Beal, P.A. Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2. Nucleic Acids Res. 2015, 43, 1123–1132.Phelps, K.J.; Tran, K.; Eifler, T.; Erickson, A.I.; Fisher, A.J.; Beal, P.A. Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2. Nucleic Acids Res. 2015, 43, 1123–1132, doi:10.1093/nar/gku1345.
- Severi, F.; Conticello, S.G. Flow-cytometric visualization of C>U mRNA editing reveals the dynamics of the process in live cells. RNA Biol. 2015, 12, 389–397.Severi, F.; Conticello, S.G. Flow-cytometric visualization of C>U mRNA editing reveals the dynamics of the process in live cells. RNA Biol. 2015, 12, 389–397, doi:10.1080/15476286.2015.1026033.
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005.Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005, doi:10.1038/nbt996.
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80.Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80, doi:10.1038/19992.
- Hsiao, Y.E.; Bahn, J.H.; Yang, Y.; Lin, X.; Tran, S.; Yang, E.W.; Quinones-Valdez, G.; Xiao, X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018, 28, 812–823.Hsiao, Y.E.; Bahn, J.H.; Yang, Y.; Lin, X.; Tran, S.; Yang, E.W.; Quinones-Valdez, G.; Xiao, X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018, 28, 812–823, doi:10.1101/gr.231209.117.
- Huang, H.; Kapeli, K.; Jin, W.; Wong, Y.P.; Arumugam, T.V.; Koh, J.H.; Srimasorn, S.; Mallilankaraman, K.; Chua, J.J.E.; Yeo, G.W.; et al. Tissue-selective restriction of RNA editing of CaV1.3 by splicing factor SRSF9. Nucleic Acids Res. 2018, 46, 7323–7338.Huang, H.; Kapeli, K.; Jin, W.; Wong, Y.P.; Arumugam, T.V.; Koh, J.H.; Srimasorn, S.; Mallilankaraman, K.; Chua, J.J.E.; Yeo, G.W.; et al. Tissue-selective restriction of RNA editing of CaV1.3 by splicing factor SRSF9. Nucleic Acids Res. 2018, 46, 7323–7338, doi:10.1093/nar/gky348.
- Tang, S.J.; Shen, H.; An, O.; Hong, H.; Li, J.; Song, Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Bellido Molias, F.; et al. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020, 11, 799.Tang, S.J.; Shen, H.; An, O.; Hong, H.; Li, J.; Song, Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Bellido Molias, F.; et al. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020, 11, 799, doi:10.1038/s41467-020-14621-5.
- Agranat, L.; Sperling, J.; Sperling, R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010, 7, 253–262.Agranat, L.; Sperling, J.; Sperling, R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010, 7, 253–262, doi:10.4161/rna.7.2.11568.
- Agranat-Tamir, L.; Shomron, N.; Sperling, J.; Sperling, R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014, 42, 4640–4651.Agranat-Tamir, L.; Shomron, N.; Sperling, J.; Sperling, R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014, 42, 4640–4651, doi:10.1093/nar/gkt1413.
- Mahlab-Aviv, S.; Boulos, A.; Peretz, A.R.; Eliyahu, T.; Carmel, L.; Sperling, R.; Linial, M. Small RNA sequences derived from pre-microRNAs in the supraspliceosome. Nucleic Acids Res. 2018, 46, 11014–11029.Mahlab-Aviv, S.; Boulos, A.; Peretz, A.R.; Eliyahu, T.; Carmel, L.; Sperling, R.; Linial, M. Small RNA sequences derived from pre-microRNAs in the supraspliceosome. Nucleic Acids Res. 2018, 46, 11014–11029, doi:10.1093/nar/gky791.
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150.Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150, doi:10.1038/s41586-020-2105-3.
- Azam, S.; Hou, S.; Zhu, B.; Wang, W.; Hao, T.; Bu, X.; Khan, M.; Lei, H. Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus. RNA Biol. 2019, 16, 1001–1009.Azam, S.; Hou, S.; Zhu, B.; Wang, W.; Hao, T.; Bu, X.; Khan, M.; Lei, H. Nuclear retention element recruits U1 snRNP com-ponents to restrain spliced lncRNAs in the nucleus. RNA Biol. 2019, 16, 1001–1009, doi:10.1080/15476286.2019.1620061.
- Wang, K.; Yin, C.; Du, X.; Chen, S.; Wang, J.; Zhang, L.; Wang, L.; Yu, Y.; Chi, B.; Shi, M.; et al. A U2-snRNP-independent role of SF3b in promoting mRNA export. Proc. Natl. Acad. Sci. USA 2019, 116, 7837–7846.Wang, K.; Yin, C.; Du, X.; Chen, S.; Wang, J.; Zhang, L.; Wang, L.; Yu, Y.; Chi, B.; Shi, M.; et al. A U2-snRNP-independent role of SF3b in promoting mRNA export. Proc. Natl. Acad. Sci. USA 2019, 116, 7837–7846, doi:10.1073/pnas.1818835116.
- Kim, J.S.; He, X.; Liu, J.; Duan, Z.; Kim, T.; Gerard, J.; Kim, B.; Pillai, M.M.; Lane, W.S.; Noble, W.S.; et al. Systematic proteomics of endogenous human cohesin reveals an interaction with diverse splicing factors and RNA-binding proteins required for mitotic progression. J. Biol. Chem. 2019, 294, 8760–8772.Kim, J.S.; He, X.; Liu, J.; Duan, Z.; Kim, T.; Gerard, J.; Kim, B.; Pillai, M.M.; Lane, W.S.; Noble, W.S.; et al. Systematic prote-omics of endogenous human cohesin reveals an interaction with diverse splicing factors and RNA-binding proteins re-quired for mitotic progression. J. Biol. Chem. 2019, 294, 8760–8772, doi:10.1074/jbc.RA119.007832.
