Retrotransposition of protein coding genes is RNA-based gene duplication leading to the creation of single exon nonfunctional copies. Nevertheless, over time, many of these duplicates acquire transcriptional capabilities. In human in most cases, these so-called retrogenes do not code for proteins but function as regulatory long noncoding RNAs (lncRNAs). The mechanisms by which they can regulate other genes include microRNA sponging, modulation of alternative splicing, epigenetic regulation and competition for stabilizing factors, among others.
- retrocopies
- retrogenes
- retrotransposition
- retroposition
- lncRNA
- disease
- parental gene
- host gene
- regulation
1. Introduction
Retrosequences, previously described as meaningless and biologically unimportant elements, are now recognized as evolutionarily significant, and their roles in shaping genomes, transcriptomes and proteomes have become increasingly evident [1][2][3]. This type of RNA-based gene duplicate is created through retroposition, which, together with DNA-based duplication, is known to be one of the major sources of new genes [2][4][5]. Formation of a retrocopy starts with transcription of the multiexonic parental gene (Figure 1). The mature mRNA is transported to the cytoplasm where in mammals proteins from LINE1 (Long interspersed nuclear elements 1), i.e., reverse transcriptase and endonuclease, accompanied by chaperones bind to the polyA tail. This complex is transported back to the nucleus where it anneals to the broken DNA ends and undergoes reverse transcription. Created cDNA is incorporated into new genomic surroundings. The final step includes creating short flanking repeats at insertion site, so called target site duplication (TDS). The presence of the 3′ polyA tail, and flanking sequences constitute signature of LINE-mediated retrotransposition [6][7]. These copies are regarded as “dead on arrival” pseudo(retro)genes, which usually lack introns, core promoters and other regulatory elements. Retrocopies are highly represented in placental mammals, especially primates [8]. In other genomes, Drosophila for example, the number of retroposed genes is relatively low [9][10]. In early studies of duplicated genes evolution, it was postulated that usually one of the duplicates accumulates mutations and becomes nonfunctional [11][12]. However, it occurred that “relaxed” selection and evolutionary freedom, which are characteristic of the majority of duplicates, may lead not only to pseudogenization but also to the acquisition of new functions [13][14]. Over time, two new phenomena related to functional evolution after duplication have been described: (i) neofunctionalization, where one copy acquires a new function and the other one keeps the original one [15], and (ii) subfunctionalization, where maintained function is shared between duplicated genes [16][17]. Additionally, as our and other studies showed, it is also possible that the retrogene (functional retrocopy) replaces its progenitor [18][19]. In the case of retrocopies, the first step needs to be obtaining regulatory elements, and there is growing evidence that many retrocopies gained the capability to be expressed over time [4][20][21].
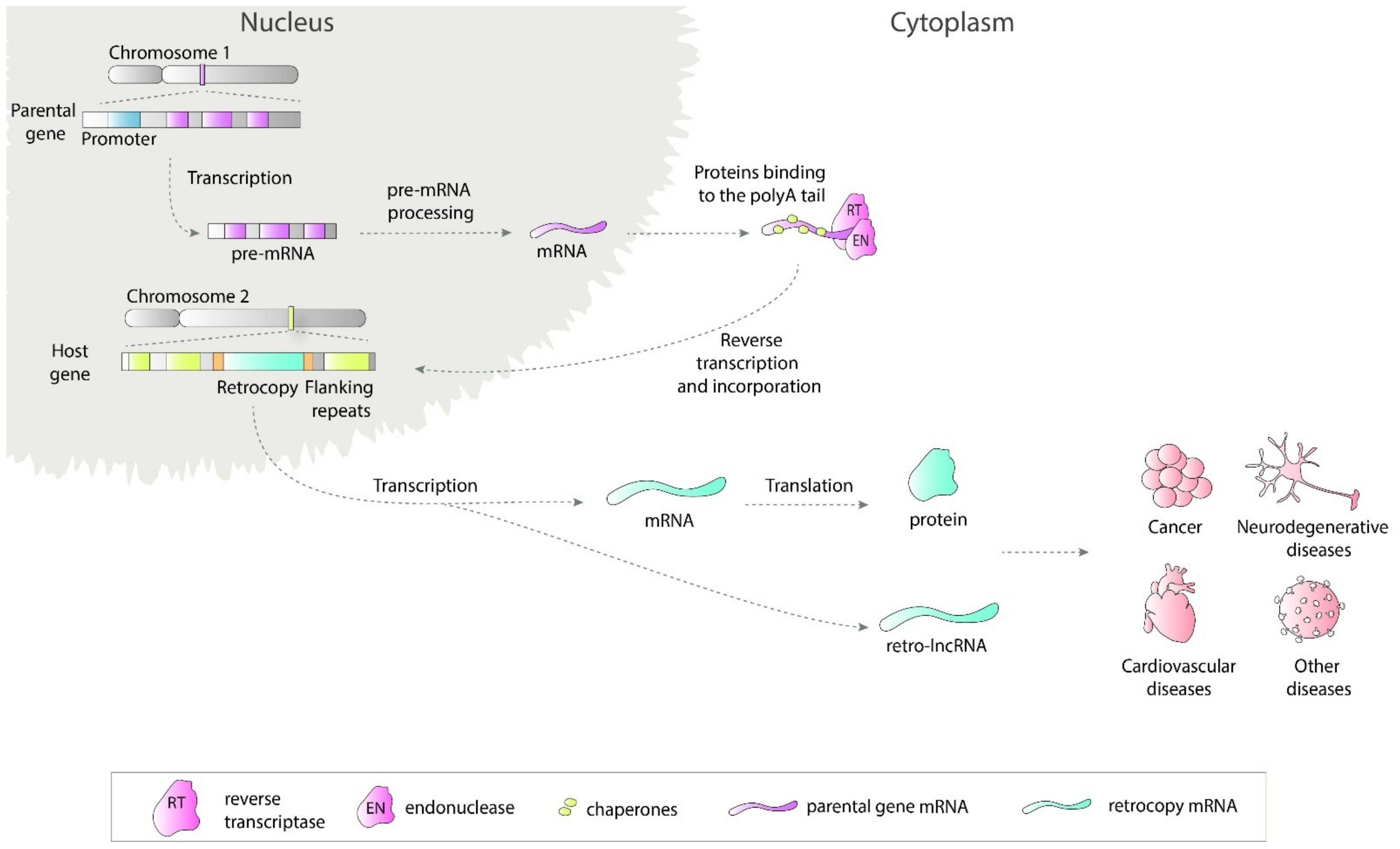
Figure 1. Retrotransposition of protein coding genes. The parental gene is transcribed and transported to the cytoplasm where LINE1-derived proteins bind to it. This complex is transported back to the nucleus and anneals to the broken DNA ends. Next, the reverse transcription process takes place and cDNA is inserted in the genome along with short flanking repeats. Transcription of created retrocopy can results in coding or non-coding RNA. Transcripts of retroposition-derived genes may be involved in pathogenesis of many human diseases.
2. Functions of Retrocopies
Regardless of being described as “junk DNA” for a long time, there are numerous examples demonstrating that retrocopies may successfully work as regulatory sequences as well as crucial protein coding genes [22][23][24]. A spectacular example of retrocopy function is the TP53 gene, a well-known tumor suppressor, and its retrocopies in elephants. Elephants have a lower-than-expected rate of cancer. It has been proposed that multiple functional retrocopies of TP53 are involved in an increased apoptotic response by compensating for the function of their progenitor [25][26]. This compensation mechanism, in turn, might underlie the cancer resistance observed in these animals. Nevertheless, in human protein coding is relatively rare among retrogenes. For example, in RetrogeneDB2 only 106 retrocopies, out of 4611, were identified as known protein coding genes, and only 847 (18%) has intact ORF (Open Reading Frame) inherited from parental gene. Interestingly, it is quite opposite in Drosophila where out of 83 identified in RetrogeneDB retrocopies, as many as 81 are annotated as known protein coding genes [27]. It was found that 256 retrocopies overlaps in the human genome with annotated lncRNAs and additional 230 may act as competing endogenous RNA since they share microRNA (miRNA) targets and have correlated expression with transcripts of 232 protein-coding genes [3]. Accumulating evidence suggests that substantial number of transcriptionally active retrocopies in human act as long noncoding RNAs (lncRNAs) [14][28]. Due to their high sequence similarity, they have a natural ability to regulate, via various mechanisms, their parental genes. Additionally, since almost 40% of retrocopies are located in introns of other genes, they possess great potential to control, as antisense transcripts, their host genes.
There are a number of ways in which retrocopies may regulate their progenitors or hosts. Retrocopies can be transcribed from the antisense strand and act as natural antisense transcripts (NATs) [29]. These NATs could be involved in multiple molecular processes, including epigenetic regulation (Figure 2A), chromatin remodeling [30], or, by forming RNA:RNA duplexes, stability control, RNA editing and processing (Figure 2B) [31]. Many retrocopies work as competing endogenous RNAs (ceRNAs), also known as microRNA sponges (Figure 2C) [15][32], while others can be a source of small RNAs [33]. Retrocopies can also compete with parental genes for other molecules, such as stabilizing factors (Figure 2D) [34] or translational machinery [35]. They may also influence the splicing of the host gene as potential factors that facilitate transcriptional interference [3][36][37][38]. The impact of retrocopies on the DNA level is also noticeable since they may be involved in nonallelic homologous recombination, resulting in the formation of chimeric transcripts (Figure 2E) [3].
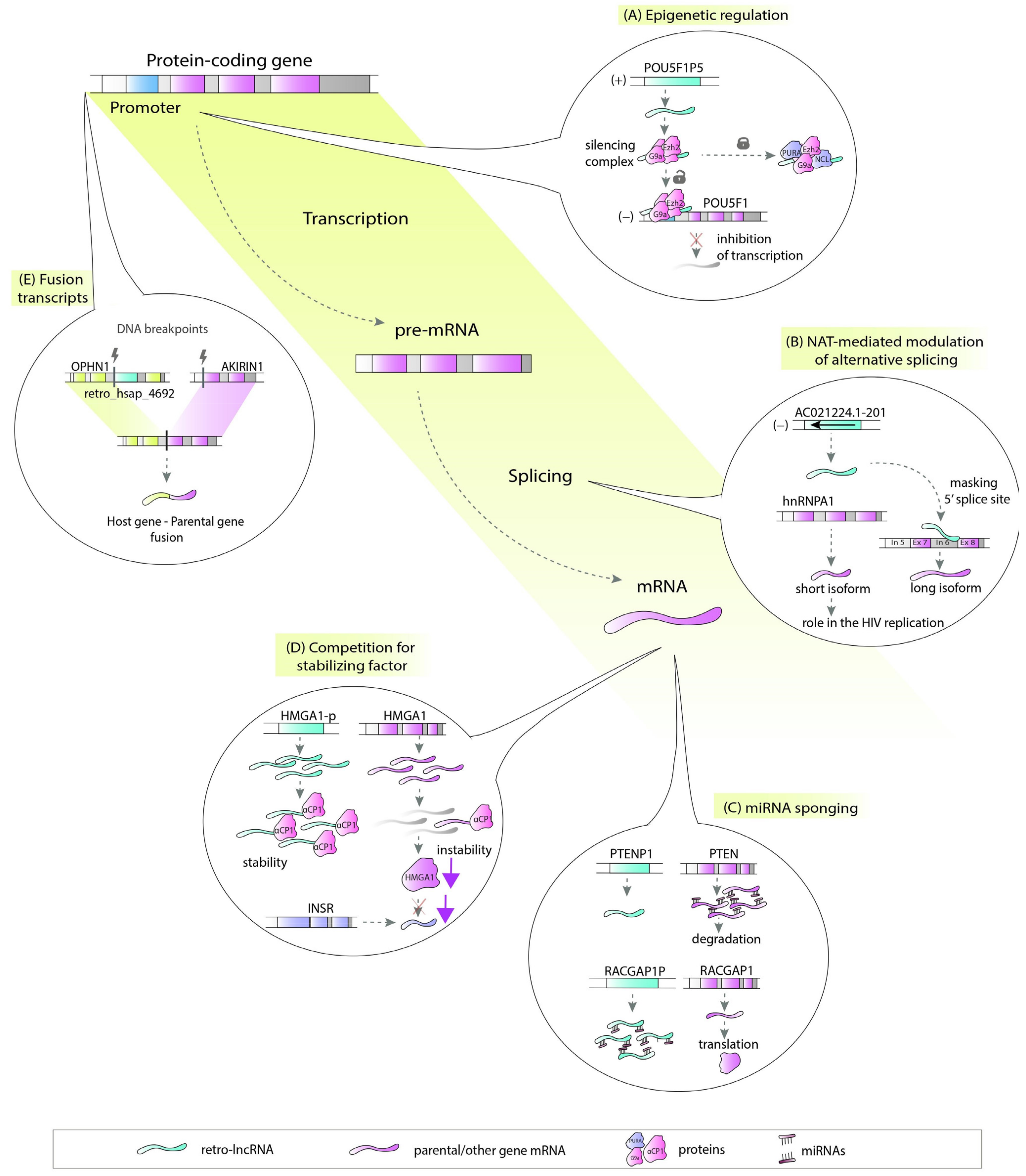
In light of the variety of possible functions, lncRNAs originating from retrocopies (retro-lncRNAs) can play a significant role in the cell regulatory machinery. This is especially important when their progenitors or host genes are critical in disease pathogenesis.
References
- Brosius, J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 1999, 238, 115–134.
- Casola, C.; Betrán, E. the genomic impact of gene retrocopies: What have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol. Evol. 2017, 9, 1351–1373.
- Kubiak, M.R.; Szcześniak, M.W.; Makałowska, I. Complex Analysis of Retroposed Genes’ Contribution to Human Genome, Proteome and Transcriptome. Genes 2020, 11, 542.
- Carelli, F.N.; Hayakawa, T.; Go, Y.; Imai, H.; Warnefors, M.; Kaessmann, H. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016, 26, 301–314.
- Betran, E.; Wang, W.; Jin, L.; Long, M. Evolution of the phosphoglycerate mutase processed gene in human and chimpanzee revealing the origin of a new primate gene. Mol. Biol. Evol. 2002, 654–663.
- Vanin, E.F. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 1985, 19, 253–272.
- Esnault, C.; Maestre, J.; Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000, 363–367.
- Mighell, A.J.; Smith, N.R.; Robinson, P.A.; Markham, A.F. Vertebrate pseudogenes. Febs Lett. 2000, 468, 109–114.
- Betran, E. Retroposed new genes out of the X in drosophila. Genome Res. 2002, 12, 1854–1859.
- Bai, Y.; Casola, C.; Feschotte, C.; Betrán, E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in drosophila. Genome Biol. 2007, 8, R11.
- Haldane, J. The part played by recurent mutation in evolution. Am. Nat. 1933, 67, 5–19.
- Fisher, R.A. The sheltering of lethals. Am. Nat. 1935, 69, 446–455.
- Nei, M. Gene duplication and nucleotide substitution in evolution. Nature 1969, 221, 40–42.
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and their mirna sponging mechanism in human cancer. Front. Cell Dev. Biol. 2020, 8, 85.
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038.
- Ohno, S. Evolution by Gene Duplication; Springer: New York, NY, USA, 1970.
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545.
- Ciomborowska, J.; Rosikiewicz, W.; Szklarczyk, D.; Makałowski, W.; Makałowska, I. “Orphan” retrogenes in the human genome. Mol. Biol. Evol. 2013, 30, 384–396.
- Krasnov, A.N. A retrocopy of a gene can functionally displace the source gene in evolution. Nucleic Acids Res. 2005, 33, 6654–6661.
- Bai, Y.; Casola, C.; Betrán, E. Evolutionary origin of regulatory regions of retrogenes in drosophila. BMC Genom. 2008.
- Sarda, S.; Hannenhalli, S. Orphan CpG islands as alternative promoters. Transcription 2018, 9, 171–176.
- Devor, E.J. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J. Hered. 2006, 97, 186–190.
- Parker, H.G.; VonHoldt, B.M.; Quignon, P.; Margulies, E.H.; Shao, S.; Mosher, D.S.; Spady, T.C.; Elkahloun, A.; Cargill, M.; Jones, P.G.; et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 2009, 325, 995–998.
- Kubiak, M.R.; Makałowska, I. Protein-Coding Genes’ Retrocopies and Their Functions. Viruses 2017, 9, 80.
- Abegglen, L.M.; Caulin, A.F.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.S.; Kiso, W.K.; Schmitt, D.L.; Waddell, P.J.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850–1860.
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.P.; Emes, R.D.; Lynch, V.J. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 2016, 5.
- Rosikiewicz, W.; Kabza, M.; Kosinski, J.G.; Ciomborowska-Basheer, J.; Kubiak, M.R.; Makalowska, I. RetrogeneDB-a database of plant and animal retrocopies. Database J. Biol. Databases Curation 2017, 2017.
- Glenfield, C.; McLysaght, A. pseudogenes provide evolutionary evidence for the competitive endogenous RNA hypothesis. Mol. Biol. Evol. 2018.
- Bryzghalov, O.; Szcześniak, M.W.; Makałowska, I. Retroposition as a source of antisense long non-coding RNAs with possible regulatory functions. Acta Biochim. Pol. 2016, 63, 825–833.
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.-O.; Corcoran, M.; Grandér, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440–446.
- Szcześniak, M.W.; Makałowska, I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS ONE 2016, 11, e0150353.
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39.
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. Mirbase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144.
- Chiefari, E.; Iiritano, S.; Paonessa, F.; Le Pera, I.; Arcidiacono, B.; Filocamo, M.; Foti, D.; Liebhaber, S.A.; Brunetti, A. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat. Commun. 2010, 1, 40.
- Bier, A.; Oviedo-Landaverde, I.; Zhao, J.; Mamane, Y.; Kandouz, M.; Batist, G. Connexin43 pseudogene in breast cancer cells offers a novel therapeutic target. Mol. Cancer Ther. 2009, 8, 786–793.
- Kaer, K.; Branovets, J.; Hallikma, A.; Nigumann, P.; Speek, M. Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation. PLoS ONE 2011, 6, e26099.
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference--a crash course. Trends Genet. TIG 2005, 21, 339–345.
- Long, M.; Langley, C.H. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 1993, 260, 91–95.
