Pharmacogenetics could explain the interindividual variability in the treatment used for the coronavirus disease 2019 (COVID-19) and improve patients’ outcomes with this complex disease. Relevant considerations should be taken into account in the design of the pharmacogenetic studies for drugs used in COVID-19.
- COVID-19
- pharmacogenetics
- CYP3A4
- CYP2D6
- ABCB1
- NR1I2
1. Introduction
Coronavirus disease 2019 (COVID-19) is a complex disorder affecting several organ systems caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In most cases, this disease clinically manifests with self-limiting mild-to-moderate symptoms of an upper respiratory tract infection, as well as general symptoms such as myalgia and fatigue. In severely affected patients, an uncontrolled immune response occurs, leading to an increase of pro-inflammatory cytokines and chemokines, and hospital and ICU care are required [1]. In severe COVID-19 patients, complications such as acute kidney injury, renal failure, myocardial injury, liver dysfunction, blood leukocyte abnormalities, septic shock, and disseminated intravascular coagulation have been described [2].
The mortality rate of COVID-19 varies among countries and the clinical conditions of the patients. The higher death rates have been associated with age, male gender, ICU care requirements, obesity, and chronic diseases (mainly oncologic, cardiovascular, metabolic, and neurodegenerative diseases) [3].
To date, there is a lack of a completely effective drug to treat COVID-19. The available treatment of COVID-19 is mainly symptomatic and is based on disease severity; however, several antimicrobials have been used for the disease treatment. The use of antiviral agents (remdesivir, lopinavir/ritonavir, oseltamivir), antibiotics (azithromycin), antiparasitics (chloroquine, hydroxychloroquine, ivermectin), and corticosteroids (dexamethasone) have been reported in the literature [4].
The evidence of the COVID-19 treatments’ effectiveness and recommendations for prescribing remains controversial, and information from clinical trials is continuously generated. Nevertheless, a lack of drug response or the occurrence of adverse drug reactions (ADR) in specific patients has been observed. For instance, some patients with COVID-19 treated with lopinavir/ritonavir have presented diarrhea, while others reported nausea and vomiting [5].
In this sense, pharmacogenetics could explain the inter-individual variability on drug response based on the genetic of COVID-19 patients [6]. Variants in genes encoding drug-metabolizing enzymes, transporters, or receptors have been reported, and they could provide the insight to achieve a personalized therapy leading to a better outcome of this emerged disease (Figure 1) [7,8,9][7][8][9]; nevertheless, several disease-related issues must be carefully reviewed in the pharmacogenetic study of COVID-19 treatment.
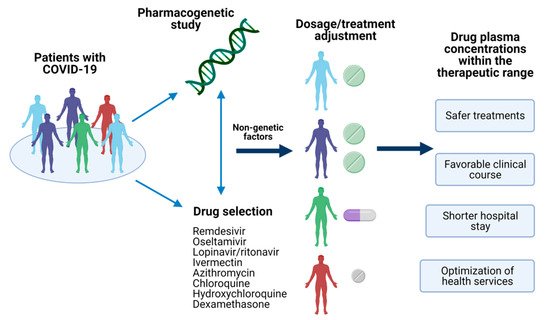
Figure 1. Pharmacogenetics of COVID-19 treatment. The identification of pharmacogenetic biomarkers of relevance in drugs used for COVID-19 treatment and nongenetic factors could provide the information to make dosages adjustments or the selection of the optimal treatment for the patient. Achieving a personalized therapy would assure drug plasma concentrations within the therapeutic range, leading to several advantages in the disease’s clinical outcome. Created with BioRender.com, accessed on 19 February 2021.
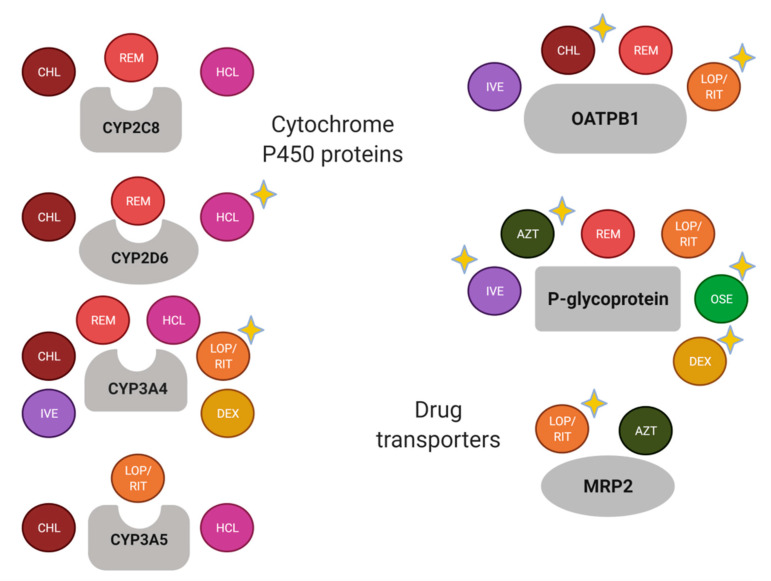
Currently, several pharmacogenetic biomarkers have been reported for drugs that have been used for COVID-19 treatment (Figure 2).
2. Considerations in Pharmacogenetic Studies of COVID-19 Treatment
COVID-19 has been a challenge for worldwide science and public health. Treatment recommendations with available drugs for this emerging disease have been established and adjusted since the beginning of the pandemic. Proposed drugs have been used for other infectious and non-infectious diseases, and therefore there is a background for the design of pharmacogenetic studies related to COVID-19 treatment [7]. However, other genetic and nongenetic factors should be taken into account, particularly for the treatment of COVID-19.
2.1. Drug-Drug Interactions
Most of the drugs used for the COVID-19 treatment are metabolized by CYP3A4 and are the substrate of P-gp and OATPB1 (
Figure 2). Nevertheless, there are well-known drug-drug interactions related to the enzyme and the transporters, which should be considered in the study of drug response variability. For instance, relevant transporter interactions of chloroquine, hydroxychloroquine, ivermectin, ritonavir, lopinavir, favipiravir, and remdesivir with the ABCB1/P-gp, ABCG2/BCRP, and ABCC1/MRP1 exporters, as well as the OATP2B1 and OATP1A2 uptake transporters, have been reported [73].
). Nevertheless, there are well-known drug-drug interactions related to the enzyme and the transporters, which should be considered in the study of drug response variability. For instance, relevant transporter interactions of chloroquine, hydroxychloroquine, ivermectin, ritonavir, lopinavir, favipiravir, and remdesivir with the ABCB1/P-gp, ABCG2/BCRP, and ABCC1/MRP1 exporters, as well as the OATP2B1 and OATP1A2 uptake transporters, have been reported [10].
Schematic summary of drug-metabolizing enzymes and transporters of drugs used for the COVID-19 treatment. A yellow-star has been added in drugs with relevant pharmacogenetic knowledge considering if the enzyme or transporter meets the following: (1) it is considered as a Pharmacogenomic Biomarker according to the FDA (
Figure 2. Schematic summary of drug-metabolizing enzymes and transporters of drugs used for the COVID-19 treatment. A yellow-star has been added in drugs with relevant pharmacogenetic knowledge considering if the enzyme or transporter meets the following: (1) it is considered as a Pharmacogenomic Biomarker according to the FDA (
, accessed on 19 February 2021); (2) it is included as pharmacogene variant in Pharmgkb (
https://www.pharmgkb.org/, accessed on 19 February 2021); and/or (3) it has been associated with the pharmacokinetics and/or pharmacodynamics of the corresponding drug in a scientific report. AZT, azithromycin; CHL, chloroquine; DEX, dexamethasone; HCL, hydroxychloroquine; IVE, ivermectin; LOP, lopinavir; OSE, oseltamivir; REM, remdesivir; RIT, ritonavir. Created with BioRender.com, accessed on 19 February 2021.
, accessed on 19 February 2021); and/or (3) it has been associated with the pharmacokinetics and/or pharmacodynamics of the corresponding drug in a scientific report. AZT, azithromycin; CHL, chloroquine; DEX, dexamethasone; HCL, hydroxychloroquine; IVE, ivermectin; LOP, lopinavir; OSE, oseltamivir; REM, remdesivir; RIT, ritonavir. Created with BioRender.com, accessed on 19 February 2021.
As a complex disease, polypharmacy could be shared among patients with COVID-19, besides their routine treatment if they present chronic co-morbidities. In this sense, patients with COVID-19 could be treated with antimicrobial, anti-inflammatory, as well as chronic treatments (e.g., antidiabetic and antihypertensive agents), and the potential effect of the co-treatment in the drug response variability should be evaluated. In
Table 3, several drugs considered as substrates, inhibitors, and inducers of CYP3A4, ABCB1, and OATPB1 are included. As observed, commonly prescribed drugs can be involved in drug interactions, including those for the COVID-19 treatment.
1, several drugs considered as substrates, inhibitors, and inducers of CYP3A4, ABCB1, and OATPB1 are included. As observed, commonly prescribed drugs can be involved in drug interactions, including those for the COVID-19 treatment.
Table 3
Table 1. Examples of drugs reported as substrates, inhibitors and/or inducers of CYP3A4, P-glycoprotein, and OATPB1 1.
| Protein |
Protein |
Substrates |
Substrates |
Strong Inhibitors |
Strong Inhibitors |
Strong Inducers |
Strong Inducers |
|---|---|---|---|---|---|---|---|
| CYP3A4 |
CYP3A4 |
Alfentanil, avanafil, buspirone, conivaptan, darifenacin, darunavir, ebastine, everolimus, ibrutinib, lomitapide, lovastatin, midazolam, naloxegol, nisoldipine, saquinavir, simvastatin, sirolimus, tacrolimus, tipranavir, triazolam, vardenafil, budesonide, dasatinib, dronedarone, eletriptan, eplerenone, felodipine, indinavir, lurasidone, maraviroc, quetiapine, sildenafil, ticagrelor, tolvaptan |
Alfentanil, avanafil, buspirone, conivaptan, darifenacin, darunavir, ebastine, everolimus, ibrutinib, lomitapide, lovastatin, midazolam, naloxegol, nisoldipine, saquinavir, simvastatin, sirolimus, tacrolimus, tipranavir, triazolam, vardenafil, budesonide, dasatinib, dronedarone, eletriptan, eplerenone, felodipine, indinavir, lurasidone, maraviroc, quetiapine, sildenafil, ticagrelor, tolvaptan |
Boceprevir, cobicistat, danoprevir and ritonavir, elvitegravir and ritonavir, grapefruit juice, indinavir and ritonavir, itraconazole, ketoconazole, lopinavir and ritonavir, paritaprevir and ritonavir, posaconazole, ritonavir, saquinavir and ritonavir, telaprevir, tipranavir and ritonavir, telithromycin, troleandomycin, voriconazole |
Boceprevir, cobicistat, danoprevir and ritonavir, elvitegravir and ritonavir, grapefruit juice, indinavir and ritonavir, itraconazole, ketoconazole, lopinavir and ritonavir, paritaprevir and ritonavir, posaconazole, ritonavir, saquinavir and ritonavir, telaprevir, tipranavir and ritonavir, telithromycin, troleandomycin, voriconazole |
Apalutamide, carbamazepine, enzalutamide, mitotane, phenytoin, rifampin, St. John’s wort |
Apalutamide, carbamazepine, enzalutamide, mitotane, phenytoin, rifampin, St. John’s wort |
| P-gp |
P-gp |
Dabigatran etexilate, digoxin, fexofenadine |
Dabigatran etexilate, digoxin, fexofenadine |
Amiodarone, carvedilol, clarithromycin, dronedarone, itraconazole, lapatinib, lopinavir and ritonavir, propafenone, quinidine, ranolazine, ritonavir, saquinavir and ritonavir, telaprevir, tipranavir and ritonavir, verapamil |
Amiodarone, carvedilol, clarithromycin, dronedarone, itraconazole, lapatinib, lopinavir and ritonavir, propafenone, quinidine, ranolazine, ritonavir, saquinavir and ritonavir, telaprevir, tipranavir and ritonavir, verapamil |
- |
- |
| OATPB1 |
OATPB1 |
Asunaprevir, atorvastatin, bosentan, danoprevir, docetaxel, fexofenadine, glyburide, nateglinide, paclitaxel, pitavastatin, pravastatin, repaglinide, rosuvastatin, simvastatin acid |
Asunaprevir, atorvastatin, bosentan, danoprevir, docetaxel, fexofenadine, glyburide, nateglinide, paclitaxel, pitavastatin, pravastatin, repaglinide, rosuvastatin, simvastatin acid |
Atazanavir and ritonavir, clarithromycin, cyclosporine, erythromycin, gemfibrozil, lopinavir and ritonavir, rifampin, simeprevir |
Atazanavir and ritonavir, clarithromycin, cyclosporine, erythromycin, gemfibrozil, lopinavir and ritonavir, rifampin, simeprevir |
- |
- |
1
Data from Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers, US Food and Drug Administration. Available in
, accessed on 19 February 2021.
2.2. Orphane Nuclear Receptors
Regarding the inhibition or induction of enzymes and drug transporters, it is also worth mentioning that two genes encode relevant nuclear receptors that contribute to both auto-induction of drug clearance and drug–drug interactions in combined therapies. The orphan nuclear receptors PXR (pregnane X receptor, encoded by
NR1I2
) and CAR (constitutive androstane receptor, encoded by
NR1I3) are xenobiotics’ sensors that mediate drug-induced changes by increasing transcription of genes involved in drug clearance and disposition. Therefore, genetic variability in these nuclear receptors could also contribute to the drugs’ response [77,78].
) are xenobiotics’ sensors that mediate drug-induced changes by increasing transcription of genes involved in drug clearance and disposition. Therefore, genetic variability in these nuclear receptors could also contribute to the drugs’ response [11][12].
PXR is an approximately 434-amino acid, 50-kDa protein, mainly expressed in the liver and intestine. It contains an N-terminus region; a DNA binding domain consisting of two zinc fingers (amino acids 41–107); a hinge region (amino acids 107–141) and a ligand-binding domain containing the ligand-binding pocket, and a ligand-depending activation factor domain (amino acids 141–434) [79,80]. When a ligand binds to PXR, the receptor is activated, and it forms a heterodimer with 9-cis retinoic acid receptor RXR-alpha, which binds to the specific DNA region of the target genes to induce their expression [81]. PXR ligands include drugs, carcinogens, food additives, pesticides, and environmental pollutants. A wide variety of drugs that bind to PXR have been described, including antibiotics, anticancer drugs, antihypertensive, antifungal [82], and examples of these drugs are included in
PXR is an approximately 434-amino acid, 50-kDa protein, mainly expressed in the liver and intestine. It contains an N-terminus region; a DNA binding domain consisting of two zinc fingers (amino acids 41–107); a hinge region (amino acids 107–141) and a ligand-binding domain containing the ligand-binding pocket, and a ligand-depending activation factor domain (amino acids 141–434) [13][14]. When a ligand binds to PXR, the receptor is activated, and it forms a heterodimer with 9-cis retinoic acid receptor RXR-alpha, which binds to the specific DNA region of the target genes to induce their expression [15]. PXR ligands include drugs, carcinogens, food additives, pesticides, and environmental pollutants. A wide variety of drugs that bind to PXR have been described, including antibiotics, anticancer drugs, antihypertensive, antifungal [16], and examples of these drugs are included in
2.
Table 4
Table 2. Ligands and target genes of the nuclear receptor PXR [14][16][17].
| Nuclear Receptor |
Nuclear Receptor |
Drug Ligands |
Drug Ligands |
Target Genes |
Target Genes |
|---|---|---|---|---|---|
| PXR |
PXR |
Amoxicillin, ampicillin, penicillin, cefuroxime, cephalexin, cefradine, sulfamethazine, erythromycin, rifampin, tetracycline, topiramate, carbamazepine, phenytoin, valproic acid, terbinafine, griseofulvin, clotrimazole, miconazole, nifedipine, cyclophosphamide, cisplatin, docetaxel, paclitaxel, vinblastine, troglitazone, rosiglitazone, atorvastatin, simvastatin, efavirenz, nevirapine, ritonavir, omeprazole, lansoprazole |
Amoxicillin, ampicillin, penicillin, cefuroxime, cephalexin, cefradine, sulfamethazine, erythromycin, rifampin, tetracycline, topiramate, carbamazepine, phenytoin, valproic acid, terbinafine, griseofulvin, clotrimazole, miconazole, nifedipine, cyclophosphamide, cisplatin, docetaxel, paclitaxel, vinblastine, troglitazone, rosiglitazone, atorvastatin, simvastatin, efavirenz, nevirapine, ritonavir, omeprazole, lansoprazole |
CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP3A4, CES2, UGT1A1, UGT1A6, ABCB1, ABCC2 |
CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP3A4, CES2, UGT1A1, UGT1A6, ABCB1, ABCC2 |
As it has been mentioned before, the target genes include phase I and II drug-metabolizing enzymes, as well as uptake and efflux drug transporters (
Table 4). The expression of these genes can be modified when a PXR ligand binds to the receptor, but an impact of
2). The expression of these genes can be modified when a PXR ligand binds to the receptor, but an impact of
NR1I2 genotype in the enzyme and transporter induction and/or in the DNA binding has also been observed [80]. The effect of several
genotype in the enzyme and transporter induction and/or in the DNA binding has also been observed [14]. The effect of several
NR1I2 variants on different drugs’ metabolism can be found in the literature. For instance: rs3814055 in erythromycin metabolism [83]; rs1464603 and rs1464602 in midazolam clearance [84]; and, rs3814058 and rs2276707 in doxorubicin clearance [85].
variants on different drugs’ metabolism can be found in the literature. For instance: rs3814055 in erythromycin metabolism [18]; rs1464603 and rs1464602 in midazolam clearance [19]; and, rs3814058 and rs2276707 in doxorubicin clearance [20].
CAR is encoded by the
NR1I3 located in chromosome 1, and it consists of nine exons. The exons 2, 3, and 4 determine the DNA binding domain, while the ligand-binding domain is encoded by the sequence of DNA comprised between the end of the exon 4 and the beginning of the 9 [78]. CAR forms a heterodimer with retinoid X receptor that binds to retinoic acid response elements and activates target genes. It shares with PXR significant cross-talk in both target gene recognition by binding to the similar xenobiotic responsive elements in their target gene promoters and accommodating a diverse array of xenobiotic activators. CAR target genes include
located in chromosome 1, and it consists of nine exons. The exons 2, 3, and 4 determine the DNA binding domain, while the ligand-binding domain is encoded by the sequence of DNA comprised between the end of the exon 4 and the beginning of the 9 [12]. CAR forms a heterodimer with retinoid X receptor that binds to retinoic acid response elements and activates target genes. It shares with PXR significant cross-talk in both target gene recognition by binding to the similar xenobiotic responsive elements in their target gene promoters and accommodating a diverse array of xenobiotic activators. CAR target genes include
CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP3A4, UGT1A1, ABCB1, ABCC2, ABCC3,
and
ABCC4 [86]. Although this receptor has been less studied than PXR, several genetic variants affecting the DNA and ligand-binding domains have been described [78].
[17]. Although this receptor has been less studied than PXR, several genetic variants affecting the DNA and ligand-binding domains have been described [12].
2.3. CYP450 Enzymes’ Expression in Inflammation and Infection Processes
Although inflammation and/or infection have not been commonly considered in pharmacogenetic studies, both processes are associated with decreased hepatic expression and/or activities of hepatic and extrahepatic CYP enzymes, drug metabolism, and drug transporters, resulting in a disturbance of the bioavailability of oral drugs [87,88]. Furthermore, the regulation of CYP enzymes’ expression mediated by several cytokines has been reported. In addition, mRNAs down-regulation of several CYPs by interleukin-6 have been observed in human hepatocytes [89], and this interleukin plays a crucial role in the cytokine storm of COVID-19 [90]. Moreover,
Although inflammation and/or infection have not been commonly considered in pharmacogenetic studies, both processes are associated with decreased hepatic expression and/or activities of hepatic and extrahepatic CYP enzymes, drug metabolism, and drug transporters, resulting in a disturbance of the bioavailability of oral drugs [21][22]. Furthermore, the regulation of CYP enzymes’ expression mediated by several cytokines has been reported. In addition, mRNAs down-regulation of several CYPs by interleukin-6 have been observed in human hepatocytes [23], and this interleukin plays a crucial role in the cytokine storm of COVID-19 [24]. Moreover,
IL6
and
IL6R variants have been recently proposed as prognostic and pharmacogenetic biomarkers of COVID-19, mainly for monoclonal antibodies targeting IL6 and IL6R [91].
variants have been recently proposed as prognostic and pharmacogenetic biomarkers of COVID-19, mainly for monoclonal antibodies targeting IL6 and IL6R [25].
Standard dosages in patients with the infectious and inflammatory process, as COVID-19, could increase exposure to the drugs, resulting in a higher possibility of ADR incidence. Simultaneously, for pro-drugs activated by metabolism, the impairment of P450 activities due to inflammation could reduce their therapeutic efficacy. In this sense, inflammatory markers and genes related to the immune response could also be considered in evaluating the inter-individual variability in the responses to COVID-19 treatment.
3. Discussion
Several pharmacogenetic biomarkers related to the metabolic pathway of drugs used for COVID-19 treatment have been described in the present review. In agreement with previous reports [7,8,9], there are variants in
Several pharmacogenetic biomarkers related to the metabolic pathway of drugs used for COVID-19 treatment have been described in the present review. In agreement with previous reports [7][8][9], there are variants in
CYP2C8
,
CYP2D6
,
CYP3A4
,
CYP3A5
,
SLCO2B1
,
ABCB1
,
ABCC2
,
CES1
, and
G6PD
that could help to improve the clinical outcome of the COVID-19. The scientific evidence supports the study of variants in
CYP2D6, CYP3A4
,
SLCO2B1
,
ABCB1,
and
ABCC2
with the response to specific drugs (
Figure 2). Nevertheless, the remaining pharmacogenes should not be discarded because the recommendations and association results are substrate-depending [92], and there is an important influence of the ethnic origin of the studied population [93].
). Nevertheless, the remaining pharmacogenes should not be discarded because the recommendations and association results are substrate-depending [26], and there is an important influence of the ethnic origin of the studied population [27].
In addition, it is necessary to consider that the drug response results from the gene-environment interaction, in which nongenetic factors (e.g., age, gender, co-treatment, disease severity) must be considered in the pharmacogenetic studies [94]. In this sense, drug-drug interactions and the inflammation and infection processes in COVID-19 could represent relevant sources of therapeutic failures and/or drug toxicity; thus the pharmacogenetic studies should identify the impact of these factors in drug response to determine the precise influence of genetic variants in the COVID-19 treatment [95,96].
In addition, it is necessary to consider that the drug response results from the gene-environment interaction, in which nongenetic factors (e.g., age, gender, co-treatment, disease severity) must be considered in the pharmacogenetic studies [28]. In this sense, drug-drug interactions and the inflammation and infection processes in COVID-19 could represent relevant sources of therapeutic failures and/or drug toxicity; thus the pharmacogenetic studies should identify the impact of these factors in drug response to determine the precise influence of genetic variants in the COVID-19 treatment [29][30].
4. Conclusions
Pharmacogenetics provides insight for the treatment improvement of several diseases, particularly for those treated with drugs presenting a wide inter-individual variability. Several pharmacogenetic markers could be evaluated in the COVID-19 treatment, which is currently based on antivirals, antibiotics, antiparasitics, and/or anti-inflammatory drugs previously used for other infectious and non-infectious diseases. Nevertheless, there are characteristics of the complex disease and the pharmacogenetic biomarkers that should be considered in the design of pharmacogenetic studies of COVID-19. Prospective studies, preferably, besides adequate control of the disease and treatment variables, could lead to valid results for treatment recommendations on the way to personalized therapy in COVID-19.
Besides, future pharmacogenetic markers should be identified for the drugs designed explicitly for the SARS-CoV-2, in which the evaluation of the virus variants in the drug response is warranted.
References
- Roberts, C.M.; Levi, M.; McKee, M.; Schilling, R.; Lim, W.S.; Grocott, M.P.W. COVID-19: A complex multisystem disorder. Br. J. Anaesth. 2020, 125, 238–242.
- Zheng, K.I.; Feng, G.; Liu, W.Y.; Targher, G.; Byrne, C.D.; Zheng, M.H. Extrapulmonary complications of COVID-19: A multisystem disease? J. Med. Virol. 2020, 93, 323–335.
- De Larochelambert, Q.; Marc, A.; Antero, J.; Le Bourg, E.; Toussaint, J.-F. Covid-19 mortality: A matter of vulnerability among nations facing limited margins of adaptation. Front. Public Health 2020, 8, 604339.
- COVID-19 Treatment Guidelines. Available online: (accessed on 19 December 2020).
- Gregoire, M.; Le Turnier, P.; Gaborit, B.J.; Veyrac, G.; Lecomte, R.; Boutoille, D.; Canet, E.; Imbert, B.-M.; Bellouard, R.; Raffi, F. Lopinavir pharmacokinetics in COVID-19 patients. J. Antimicrob. Chemother. 2020, 75, 2702–2704.
- Bishop, J.R. Pharmacogenetics. In Handbook of Clinical Neurology; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 147, pp. 59–73.
- Takahashi, T.; Luzum, J.A.; Nicol, M.R.; Jacobson, P.A. Pharmacogenomics of COVID-19 therapies. Genom. Med. 2020, 5, 35.
- Babayeva, M.; Loewy, Z. Repurposing drugs for COVID-19: Pharmacokinetics and pharmacogenomics of chloroquine and hydroxychloroquine. Pharmgenom. Pers. Med. 2020, 13, 531–542.
- Zubiaur, P.; Koller, D.; Saiz-Rodríguez, M.; Navares-Gómez, M.; Abad-Santos, F. Important pharmacogenetic information for drugs prescribed during the SARS-CoV-2 infection (COVID-19). Clin. Transl. Sci. 2020, 13, 1023–1033.
- Telbisz, Á.; Ambrus, C.; Mózner, O.; Szabó, E.; Várady, G.; Bakos, É.; Sarkadi, B.; Özvegy-Laczka, C. Interactions of anti-COVID-19 drug candidates with multispecific ABC and OATP drug transporters. bioRxiv 2020.
- Lamba, J.; Lamba, V.; Strom, S.; Venkataramanan, R.; Schuetz, E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab. Dispos. 2008, 36, 169–181.
- Lamba, J.; Lamba, V.; Schuetz, E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr. Drug Metab. 2005, 6, 369–383.
- Carnahan, V.; Redinbo, M. Structure and function of the human nuclear xenobiotic receptor PXR. Curr. Drug Metab. 2005, 6, 357–367.
- Oshiro, C. Very Important Pharmacogene: NR1I2. Available online: https://www.pharmgkb.org/vip/PA166170351 (accessed on 24 January 2021).
- Lehmann, J.; McKee, D.; Watson, M.; Willson, T.; Moore, J.; Kliewer, S. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Investig. 1998, 102, 1016–1023.
- Pavek, P. Pregnane X Receptor (PXR)-mediated gene repression and cross-talk of PXR with other nuclear receptors via coactivator interactions. Front. Pharmacol. 2016, 7, 456.
- Tolson, A.H.; Wang, H. Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv. Drug Deliv. Rev. 2010, 62, 1238–1249.
- Zhang, J.; Kuehl, P.; Green, E.D.; Touchman, J.W.; Watkins, P.B.; Daly, A.; Hall, S.D.; Maurel, P.; Relling, M.; Brimer, C.; et al. The human pregnane X receptor: Genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 2001, 11, 555–572.
- He, P.; Court, M.; Greenblatt, D.; von Moltke, L.L. Human pregnane X receptor: Genetic polymorphisms, alternative mRNA splice variants, and cytochrome P450 3A metabolic activity. J. Clin. Pharmacol. 2006, 46, 1356–1369.
- Sandanaraj, E.; Lal, S.; Selvarajan, V.; Ooi, L.; Wong, Z.; Wong, N.; Ang, P.; Lee, E.; Chowbay, B. PXR pharmacogenetics: Association of haplotypes with hepatic CYP3A4 and ABCB1 messenger RNA expression and doxorubicin clearance in Asian breast cancer patients. Clin. Cancer Res. 2008, 14, 7116–7126.
- Morgan, E.T. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin. Pharmacol. Ther. 2009, 85, 434–438.
- Stavropoulou, E.; Pircalabioru, G.G.; Bezirtzoglou, E. The role of cytochromes P450 in infection. Front. Immunol. 2018, 9, 31.
- Aitken, A.E.; Morgan, E.T. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab. Dispos. 2007, 35, 1687–1693.
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Sharma, G.; Lee, S.; Agoramoorthy, G. COVID-19: Consider IL-6 receptor antagonist for the therapy of cytokine storm syndrome in SARS-CoV-2 infected patients. J. Med. Virol. 2020.
- Strafella, C.; Caputo, V.; Termine, A.; Barati, S.; Caltagirone, C.; Giardina, E.; Cascella, R. Investigation of genetic variations of IL6 and IL6r as potential prognostic and pharmacogenetics biomarkers: Implications for covid-19 and neuroinflammatory disorders. Life 2020, 10, 351.
- Deenen, M.J.; Cats, A.; Beijnen, J.H.; Schellens, J.H.M. Part 1: Background, methodology, and clinical adoption of pharmacogenetics. Oncologist 2011, 16, 811–819.
- Mizzi, C.; Dalabira, E.; Kumuthini, J.; Dzimiri, N.; Balogh, I.; Başak, N.; Böhm, R.; Borg, J.; Borgiani, P.; Bozina, N.; et al. A European spectrum of pharmacogenomic biomarkers: Implications for clinical pharmacogenomics. PLoS ONE 2016, 11, e0162866.
- Smit, R.A.J.; Noordam, R.; le Cessie, S.; Trompet, S.; Jukema, J.W. A critical appraisal of pharmacogenetic inference. Clin. Genet. 2018, 93, 498–507.
- Shah, R.R. Pharmacogenetics and precision medicine: Is inflammation a covert threat to effective genotype-based therapy? Ther. Adv. Drug Saf. 2017, 8, 267–272.
- Via, M.; Tcheurekdjian, H.; Burchard, E.G. Role of interactions in pharmacogenetic studies: Leukotrienes in asthma. Pharmacogenomics 2013, 14, 923–929.