Minor splicing plays an important role in vertebrate development. Zrsr1 and Zrsr2 paralog genes have essential roles in alternative splicing, mainly participating in the recognition of minor (U12) introns. Mice embryos with mutations in both splicing factors stopped developing mainly between the 2- and 4-cell stages, just after zygotic gene activation. RNA-seq analysis of Zrsr1/2mu 2-cell embryos showed altered gene and isoform expression of thousands of genes enriched in essential gene ontology terms and biological pathways related to ribosome, RNA transport, spliceosome, and essential zygotic gene activation steps. Alternative splicing of both U2 and U12 intron-containing genes was altered related to cell cycle and mitotic nuclear division. Zrsr1 and Zrsr2 were also required for the conversion of mouse-induced pluripotent stem cells into 2C-like cells. Zrsr1 and Zrsr2 emerge as necessary for zygotic gene activation and the conversion of induced pluripotent stem cells into 2C-like cells.
- Zrsr1
- Zrsr2
- U12 introns
- zygotic gene activation
- 2C-like cell
Introduction
Alternative splicing (AS) is an essential co- and post-transcriptional process through which multiple transcripts are generated from a single gene. There are two splicing machineries: the major class or U2-dependent spliceosome, which removes the majority of introns (U2-type intron); and the minor class or U12-dependent spliceosome, which removes U12-type introns. U12-type introns represent around 0.4% of all introns in the genome
Alternative splicing (AS) is an essential co- and post-transcriptional process through which multiple transcripts are generated from a single gene. There are two splicing machineries: the major class or U2-dependent spliceosome, which removes the majority of introns (U2-type intron); and the minor class or U12-dependent spliceosome, which removes U12-type introns. U12-type introns represent around 0.4% of all introns in the genome[1], and are non-randomly distributed across the genome. Despite their scarce abundance, minor introns are highly conserved across distantly related eukaryotic taxa, indicating their common evolutionary origin
, and are non-randomly distributed across the genome. Despite their scarce abundance, minor introns are highly conserved across distantly related eukaryotic taxa, indicating their common evolutionary origin[2][3]. Minor splicing has been by far less studied than canonical splicing, and although at first it was described as a cytoplasmatic mechanism
. Minor splicing has been by far less studied than canonical splicing, and although at first it was described as a cytoplasmatic mechanism[4][5], several studies have refuted this theory
, several studies have refuted this theory[6][7][8]. However, some level of cytoplasmatic splicing has been described in strongly compartmentalized cells, such as neurons or megakaryocytes
. However, some level of cytoplasmatic splicing has been described in strongly compartmentalized cells, such as neurons or megakaryocytes[9], but its function remains unclear.
Further, genes with U12 introns are over-represented in functions and pathways related to development, such as RNA processing, DNA replication, or cell cycle
, but its function remains unclear.[10][11], and they show tissue-dependent intron retention
, and they show tissue-dependent intron retention[12]. Several mutations of the minor spliceosome have been associated with multiple diseases, including developmental disorders
. Several mutations of the minor spliceosome have been associated with multiple diseases, including developmental disorders[10][13][14][15][16], neurodegeneration
, neurodegeneration[17], and cancer
, and cancer[18]. Additionally, the minor spliceosome is thought to play an important developmental role in plants
. Additionally, the minor spliceosome is thought to play an important developmental role in plants, drosophila[22], zebrafish
, zebrafish[23], and in the mouse central nervous system
, and in the mouse central nervous system, hypothalamus[26], gametogenesis
, gametogenesis[16]
, and early development[27]. Genes involved in mRNA splicing are over-represented during early preimplantation development
. Genes involved in mRNA splicing are over-represented during early preimplantation development[28], before and during zygotic gene activation (ZGA)
, before and during zygotic gene activation (ZGA)[29]. However, the specific role of minor splicing in preimplantation embryo development has not been fully elucidated.
The splicing factors ZRSR1 and ZRSR2 (encoded by the Zrsr1 and Zrsr2 genes, respectively) have been identified in all mammalian species analysed
. However, the specific role of minor splicing in preimplantation embryo development has not been fully elucidated.The splicing factors ZRSR1 and ZRSR2 (encoded by the Zrsr1 and Zrsr2 genes, respectively) have been identified in all mammalian species analysed
[16][30], and are involved in the recognition of the 3’AC dinucleotide of the AT-AC class of U12-type introns or the 3’AG of U2-type introns
, and are involved in the recognition of the 3’AC dinucleotide of the AT-AC class of U12-type introns or the 3’AG of U2-type introns[31]. Recent sequencing studies have identified frequent somatic ZRSR2 mutations in hematological malignancies, such as myelodysplastic syndrome (MDS), causing mis-splicing of U12 introns
. Recent sequencing studies have identified frequent somatic ZRSR2 mutations in hematological malignancies, such as myelodysplastic syndrome (MDS), causing mis-splicing of U12 introns[18]. However, their precise role in embryo development and function in other pluripotent cell types remains unclear. Zrsr1 is a retrotransposed copy of X-linked Zrsr2 (located on the X chromosome in all mammalian species analysed) and is paternally expressed in the placenta and some adult tissues, while the maternal copy is methylated and silent. Zrsr1 plays a key role in hematopoiesis, muscle strength, and spermatogenesis, leading its mutation to infertility
. However, their precise role in embryo development and function in other pluripotent cell types remains unclear. Zrsr1 is a retrotransposed copy of X-linked Zrsr2 (located on the X chromosome in all mammalian species analysed) and is paternally expressed in the placenta and some adult tissues, while the maternal copy is methylated and silent. Zrsr1 plays a key role in hematopoiesis, muscle strength, and spermatogenesis, leading its mutation to infertility[16].
.Results and discussion
Mutant Mice Generation and Rescue Experiments
To gain further insight into the role ofZrsr1
andZrsr2
in preimplantation development, in the present study we generatedZrsr1
andZrsr2
mutant mice, truncating their RNA recognition motif. Double mutants produced by crossing males homozygous for the Zrsr1 mutation with females homozygous for theZrsr2
mutation (hereafter namedZrsr1/2mu
) stopped developing just after ZGA. Rescue experiments in whichZrsr1
mRNA was injected into single-cellZrsr1/2mu
embryos extended the development of mutant embryos, revealing that minor splicing is essential for ZGA. A clear complementarity action between ZRSR1 and ZRSR2 during 2-cell embryo cleavage was observed, as embryos need at least oneZrsr1
wild-type (WT) allele from the father (as it is maternally imprinted and only expressed from the paternal allele) or oneZrsr2
WT allele from the mother (as onlyZrsr2
is expressed in the oocyte and the embryo needs the protein during early stages of development).
Transcriptomic Analysis and Conversion of Induced Pluripotent Stem Cells (iPSCs) to 2C-Like Cells
RNA-seq analysis of the transcriptome of
RNA-seq analysis of the transcriptome ofZrsr1/2mu and WT embryos revealed a large number of overexpressed genes in the double mutant embryos that are supposed to be degraded during early embryo development
and WT embryos revealed a large number of overexpressed genes in the double mutant embryos that are supposed to be degraded during early embryo development, suggesting that the mutations cause a blockade in the degradation of the oocyte’s maternal mRNAs. Intron retention was affected for both U2 and U12 introns, being exon skipping also altered. Differentially retained U2 introns located within U12-containing genes were located preferentially adjacent to U2 introns, suggesting an interplay between the major and minor spliceosomes, which goes in accordance with the results showed by Horiuchiet al, 2018
, 2018[16]. We also analysed the role of
. We also analysed the role of Zrsr1 and Zrsr2 in the conversion of induced pluripotent stem cells (iPSCs) to 2C-like cells (2CLC), which are a rare metastable cell population that appears at a very low frequency in mouse embryonic stem cells (mESCs) and iPSCs in culture. We discovered that both Zrsr1 and Zrsr2 are necessary for the generation of 2CLC, and contrary to what happens with the embryos,Zrsr1
andZrsr2 in the conversion of induced pluripotent stem cells (iPSCs) to 2C-like cells (2CLC), which are a rare metastable cell population that appears at a very low frequency in mouse embryonic stem cells (mESCs) and iPSCs in culture. We discovered that both
are not complementary in iPSCs, as both are required for the induction of 2CLC.Zrsr1 and
andZrsr2 are necessary for the generation of 2CLC, and contrary to what happens with the embryos,
are expressed that are critical. First,Zrsr1 and
andZrsr2 are not complementary in iPSCs, as both are required for the induction of 2CLC.
The different pleiotropic phenotypes of U12 splicing factors have made it difficult to identify unifying developmental functions of U12 intron-containing genes. There are several events common to gametogenesis and preimplantation development at the precise moment Zrsr1 and Zrsr2 are expressed that are critical. First, Zrsr1 and Zrsr2 are expressed in gametogenesis during the meiotic prophase, which is equivalent to a prolonged G2 phase in the mitotic cell cycle [
are expressed in gametogenesis during the meiotic prophase, which is equivalent to a prolonged G2 phase in the mitotic cell cycle[34]. Similarly, the G1 phase is extremely short, while G2 is very long during the second preimplantation division in mouse embryos, when the major phase of ZGA takes place [
]. Similarly, the G1 phase is extremely short, while G2 is very long during the second preimplantation division in mouse embryos, when the major phase of ZGA takes place[35] and Zrsr1 is specifically expressed. The gene Zscan4 is also expressed in oocytes and spermatocytes during the late meiotic prophase, in G2 phase ES cells and in late 2-cell stage mouse embryos [
] and Zrsr1 is specifically expressed. The gene Zscan4 is also expressed in oocytes and spermatocytes during the late meiotic prophase, in G2 phase ES cells and in late 2-cell stage mouse embryos[36]. Its expression is associated with heterochromatin organization [
]. Its expression is associated with heterochromatin organization[37]. Interestingly, the small proportion of genes with U12 introns in vertebrates is over-represented in specific G2 phase functions like DNA replication, cell cycle, and DNA damage repair. The disruption of multiple cell cycle and DNA damage repair genes would be expected to cause defects in cell differentiation. Another common event between meiosis, ZGA and iPSC conversion in 2CLC is the alteration in the structure of the nuclear membrane. In addition to nuclear break down that occurs in meiosis, nuclear lamina is rearranged during stem cell differentiation [
]. Interestingly, the small proportion of genes with U12 introns in vertebrates is over-represented in specific G2 phase functions like DNA replication, cell cycle, and DNA damage repair. The disruption of multiple cell cycle and DNA damage repair genes would be expected to cause defects in cell differentiation. Another common event between meiosis, ZGA and iPSC conversion in 2CLC is the alteration in the structure of the nuclear membrane. In addition to nuclear break down that occurs in meiosis, nuclear lamina is rearranged during stem cell differentiation[38], and the nuclear envelope breaks down after the two pronuclei undergo DNA replication, and chromosomes then associate during the first mitosis [
], and the nuclear envelope breaks down after the two pronuclei undergo DNA replication, and chromosomes then associate during the first mitosis[39].
According to our results, the Zrsr1/2
].mu
phenotype observed in embryos and iPSC could be the outcome of one of several disrupted procedures: dysregulation of U12-type intron-containing genes, the disruption of AS, and/or the altered expression of other genes. Our gene ontology (GO) analysis of U12 intron-bearing genes with alterations in intron retention in Zrsr1/2mu
embryos revealed the strong enrichment of genes related to key cellular functions such as cell cycle, mitotic nuclear division, DNA damage repair, RNA processing, etc., highlighting the importance of these genes in the 2-cell blockade. However, Zrsr1/2mu also indirectly modified the expression of many other genes reported to play an important role in embryo cleavage and iPSC conversion to 2C-like cells, some of them carrying U12 introns. For example, one important gene for reprogramming the conversion from mouse embryonic fibroblasts (MEF) to iPSC is the epithelia-splicing factor Esrp1 [
also indirectly modified the expression of many other genes reported to play an important role in embryo cleavage and iPSC conversion to 2C-like cells, some of them carrying U12 introns. For example, one important gene for reprogramming the conversion from mouse embryonic fibroblasts (MEF) to iPSC is the epithelia-splicing factor Esrp1[40], a marker of several cancers [
], a marker of several cancers[] that has an U12 intron. In our Zrsr1/2mu
embryos, Esrp1 showed AS alterations in ES and IR of the U12 intron. In the same manner as Esrp1, the genes Zrsr1 and Zrsr2 were necessary for iPSC reprogramming to the 2C-like state, but were not required for maintenance of pluripotency, as we could maintain Zrsr1 and Zrsr2 mutant iPSC lines indefinitely without loss of viability. Another gene down-regulated in the Zrsr1/2mu embryos was Dppa3 (also known as Stella), a marker of pluripotency and cell division that ensures activation of endogenous retroviruses (ERVs) [
embryos was Dppa3 (also known as Stella), a marker of pluripotency and cell division that ensures activation of endogenous retroviruses (ERVs)[42]. Cnot7 (also known as Caf-1) controls ERVs and genes specific to 2-cell embryos, and its depletion in ESCs enhances the reprogramming of 2C-like cells [
]. Cnot7 (also known as Caf-1) controls ERVs and genes specific to 2-cell embryos, and its depletion in ESCs enhances the reprogramming of 2C-like cells[]. This gene also showed AS alterations in ES and IR in Zrsr1/2mu embryos. Also, the Developmental pluripotency associated genes 2 (Dppa2) and 4 (Dppa4), which are positive regulators of 2C-like cells and ZGA transcription by directly up-regulating Dux [
embryos. Also, the Developmental pluripotency associated genes 2 (Dppa2) and 4 (Dppa4), which are positive regulators of 2C-like cells and ZGA transcription by directly up-regulating Dux[], were down regulated in Zrsr1/2mu embryos. Other genes showing high intron retention in mutant embryos were Srsf10 and Cul1. Srsf10 is required to implement DNA damage-induced splicing shifts in other transcripts involved in apoptosis, cell-cycle control and DNA repair, indicating that SRSF10 connects DNA damage to the alternative splicing of transcripts that determine cell fate [
embryos. Other genes showing high intron retention in mutant embryos were Srsf10 and Cul1. Srsf10 is required to implement DNA damage-induced splicing shifts in other transcripts involved in apoptosis, cell-cycle control and DNA repair, indicating that SRSF10 connects DNA damage to the alternative splicing of transcripts that determine cell fate[45]. Cul1 is the major scaffold of the SCF complex (Skp, Cullin, F-box), an E3 ubiquitin ligase that contributes to the regulation of many cell processes, including proliferation, differentiation and death, by targeting its substrate proteins for degradation via the ubiquitin-proteasome system [
]. Cul1 is the major scaffold of the SCF complex (Skp, Cullin, F-box), an E3 ubiquitin ligase that contributes to the regulation of many cell processes, including proliferation, differentiation and death, by targeting its substrate proteins for degradation via the ubiquitin-proteasome system[46].
].Conclusions
This comprehensive analysis of Zrsr1/2
This comprehensive analysis of Zrsr1/2mu mouse embryos reveals a new role of paternal
mouse embryos reveals a new role of paternalZrsr1 and maternal
and maternalZrsr2 as components of the ZGA mechanism in the embryo, as mutation of either
as components of the ZGA mechanism in the embryo, as a mutation of eitherZrsr1 or
orZrsr2 blocked the reprogramming of iPSCs towards 2CLC (Figure 1). Accordingly,
blocked the reprograming of iPSCs towards 2CLC (Figure 1). Accordingly,Zrsr1 and Zrsr2 emerge as essential for stem cell differentiation during gametogenesis
and Zrsr2 emerge as essential for stem cell differentiation during gametogenesis[16], totipotent zygote differentiation (ZGA) and reprogramming of iPSCs towards 2CLC, suggesting a critical role of minor splicing in the stem cell reprogramming process.
, totipotent zygote differentiation (ZGA), and reprogramming of iPSCs towards 2CLC, suggesting a critical role of minor splicing in the stem cell reprogramming process.
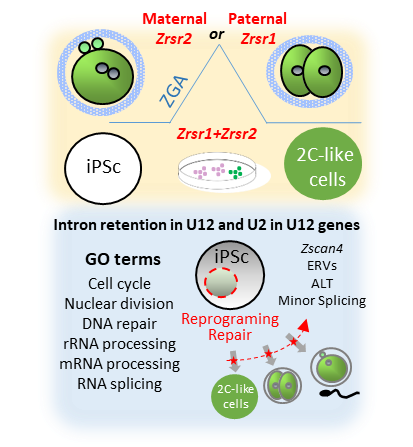
Figure 1.
Synopsis of phenotypes produced byZrsr1
andZrsr2
mutations.
Upper panel shows that mutation of maternalZrsr2
and paternalZrsr1
blocked early embryo development during zygotic gene activation (ZGA) and increased intron retention in U2- and U12-containing genes; and that mutation of eitherZrsr1
orZrsr2 led to a blockade of induced pluripotent stem cell (iPSC) conversion to 2C-like cells. The bottom panel shows gene ontology (GO) enrichment terms related to these phenotypes, indicating that disturbed U12-containing genes are critical for cell division, differentiation, and DNA damage response.
led to a blockade of induced pluripotent stem cell (iPSC) conversion to 2C-like cells. The bottom panel shows gene ontology (GO) enrichment terms related to these phenotypes, indicating that disturbed U12-containing genes are critical for cell division, differentiation, and DNA damage response.
References
- Christopher B. Burge; Richard Padgett; P A Sharp; Evolutionary Fates and Origins of U12-Type Introns. Molecular Cell 1998, 2, 773-785, 10.1016/s1097-2765(00)80292-0.
- Anthony G. Russell; J. Michael Charette; David F. Spencer; Michael W. Gray; An early evolutionary origin for the minor spliceosome. Nature 2006, 443, 863-866, 10.1038/nature05228.
- Sebastian Canzler; T Samuelsson; U12 type introns were lost at multiple occasions during evolution. BMC Genomics 2010, 11, 106-106, 10.1186/1471-2164-11-106.
- Harald König; Nathalie Matter; Rüdiger Bader; Wilko Thiele; Ferenc Müller; Splicing Segregation: The Minor Spliceosome Acts outside the Nucleus and Controls Cell Proliferation. Cell 2007, 131, 718-729, 10.1016/j.cell.2007.09.043.
- Richard Robinson; Minor spliceosome, major surprise: it's cytoplasmic. Journal of Cell Biology 2007, 179, 1086-1086, 10.1083/jcb.1796rr1.
- Kyle Friend; Nikolay G. Kolev; Mei-Di Shu; Joan A. Steitz; Minor-class splicing occurs in the nucleus of the Xenopus oocyte. RNA 2008, 14, 1459-1462, 10.1261/rna.1119708.
- Heli K. J. Pessa; Cindy L. Will; Xiaojuan Meng; Claudia Schneider; Nicholas Watkins; Nina Maria Perälä; Mariann Nymark; Janne Turunen; Reinhard Lührmann; Mikko J. Frilander; et al. Minor spliceosome components are predominantly localized in the nucleus. Proceedings of the National Academy of Sciences 2008, 105, 8655-8660, 10.1073/pnas.0803646105.
- Jarnail Singh; Richard Padgett; Rates of in situ transcription and splicing in large human genes. Nature Structural & Molecular Biology 2009, 16, 1128-33, 10.1038/nsmb.1666.
- Peter T. Buckley; Mugdha Khaladkar; Junhyong Kim; James Eberwine; Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdisciplinary Reviews: RNA 2013, 5, 223-230, 10.1002/wrna.1203.
- Daniele Merico; Maian Roifman; Ulrich Braunschweig; Ryan K. C. Yuen; Roumiana Alexandrova; Andrea Bates; Brenda Reid; Thomas Nalpathamkalam; ZhuoZhi Wang; Bhooma Thiruvahindrapuram; et al.Paul E. GrayAlyson KakakiosJane PeakeStephanie HogarthDavid MansonRaymond BuncicSergio PereiraJo-Anne HerbrickBenjamin J. BlencoweChaim M. RoifmanStephen W. Scherer Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nature Communications 2015, 6, 8718, 10.1038/ncomms9718.
- Janne Turunen; Elina Niemelä; Bhupendra Verma; Mikko J. Frilander; The significant other: splicing by the minor spliceosome. Wiley Interdisciplinary Reviews: RNA 2012, 4, 61-76, 10.1002/wrna.1141.
- Anouk M Olthof; Katery C. Hyatt; Rahul N Kanadia; Minor intron splicing revisited: identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns. BMC Genomics 2019, 20, 686-19, 10.1186/s12864-019-6046-x.
- Huiling He; Sandya Liyanarachchi; Keiko Akagi; Rebecca Nagy; Jingfeng Li; Rosemary C. Dietrich; Wei Li; Nikhil Sebastian; Bernard Wen; Baozhong Xin; et al.Jarnail SinghPearlly YanHansjuerg AlderEric HaanDagmar WieczorekBeate AlbrechtErik G. PuffenbergerHeng WangJudith A. WestmanRichard A. PadgettDavid E. SymerAlbert De La Chapelle Mutations in U4atac snRNA, a Component of the Minor Spliceosome, in the Developmental Disorder MOPD I. Science 2011, 332, 238-240, 10.1126/science.1200587.
- Mahmoud Fawzi Elsaid; Nader Chalhoub; Tawfeg Ben‐Omran; Pankaj Kumar; Hussein Kamel; Khalid Ibrahim; Yasmin Mohamoud; Eman Al‐Dous; Iman Al-Azwani; Joel A. Malek; et al.Karsten SuhreM. Elizabeth RossAlice Abdel Aleem Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Annals of Neurology 2017, 81, 68-78, 10.1002/ana.24826.
- Patrick Edery; Charles Marcaillou; Mourad Sahbatou; Audrey Labalme; Joelle Chastang; Renaud Touraine; Emmanuel Tubacher; Faiza Senni; Michael B. Bober; Sheela Nampoothiri; et al.Pierre-Simon JoukElisabeth SteichenSiren BerlandAnnick ToutainCarol WiseDamien SanlavilleFrancis RousseauFrançoise Clerget-DarpouxA. L. Leutenegger Association of TALS Developmental Disorder with Defect in Minor Splicing Component U4atac snRNA. Science 2011, 332, 240-243, 10.1126/science.1202205.
- Keiko Horiuchi; Serafín Pérez-Cerezales; Panagiotis Papasaikas; Priscila Ramos-Ibeas; Angela Patricia Lopez-Cardona; Ricardo Laguna-Barraza; Noelia Fonseca Balvís; Eva Pericuesta; Raúl Fernández-González; Benjamin Planells; et al.Alberto Viera1José Ángel SujaPablo Juan RossFrancisco AlenLaura OrioFernando Rodríguez De FonsecaBelen PintadoJuan ValcarcelAlfonso Gutierrez-Adan Impaired Spermatogenesis, Muscle, and Erythrocyte Function in U12 Intron Splicing-Defective Zrsr1 Mutant Mice. Cell Reports 2018, 23, 143-155, 10.1016/j.celrep.2018.03.028.
- Thomas Doktor; Yimin Hua; Henriette Skovgaard Andersen; Sabrina Brøner; Ying Hsiu Liu; Anna Wieckowska; Maja Dembic; Gitte Hoffmann Bruun; Adrian R. Krainer; Brage Storstein Andresen; et al. RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns.. Nucleic Acids Research 2016, 45, 395-416, 10.1093/nar/gkw731.
- Vikas Madan; Deepika Kanojia; Jia Li; Ryoko Okamoto; Aiko Sato-Otsubo; Alexander Kohlmann; Masashi Sanada; Vera Grossmann; Janani Sundaresan; Yuichi Shiraishi; et al.Satoru MiyanoFelicitas TholArnold GanserHenry YangTorsten HaferlachSeishi OgawaH. Phillip Koeffler Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome.. Nature Communications 2015, 6, 6042-6042, 10.1038/ncomms7042.
- Tao Xu; Bo Mi Kim; Kyung Jin Kwak; Hyun Ju Jung; Hunseung Kang; The Arabidopsis homolog of human minor spliceosomal protein U11-48K plays a crucial role in U12 intron splicing and plant development.. Journal of Experimental Botany 2016, 67, 3397-406, 10.1093/jxb/erw158.
- Christine Gault; Federico Martin; Wenbin Mei; Fang Bai; Joseph B. Black; William Bradley Barbazuk; A. Mark Settles; Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development.. Proceedings of the National Academy of Sciences 2017, 114, E2195-E2204, 10.1073/pnas.1616173114.
- Fang Bai; Jacob Corll; Donya N. Shodja; Ruth Davenport; Guanqiao Feng; Janaki S. Mudunkothge; Christian J. Brigolin; Federico Martin; Gertraud Spielbauer; Chi-Wah Tseung; et al.Amy E. SiebertWilliam Bradley BarbazukShailesh K LalA. Mark Settles RNA Binding Motif Protein 48 Is Required for U12 Splicing and Maize Endosperm Differentiation. The Plant Cell 2019, 31, 715-733, 10.1105/tpc.18.00754.
- Leo R. Otake; Petra Scamborova; Carl Hashimoto; Joan A. Steitz; The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila.. Molecular Cell 2002, 9, 439-446, 10.1016/s1097-2765(02)00441-0.
- Sebastian Markmiller; Nicole Cloonan; Rea M. Lardelli; Karen Doggett; M. Cristina Keightley; Yeliz Boglev; Andrew J. Trotter; Annie Y. Ng; Simon J. Wilkins; Heather Verkade; et al.Elke OberHolly A. FieldSean GrimmondGraham J. LieschkeDidier Yr StainierJoan Heath Minor class splicing shapes the zebrafish transcriptome during development.. Proceedings of the National Academy of Sciences 2014, 111, 3062-7, 10.1073/pnas.1305536111.
- Marybeth Baumgartner; Christopher Lemoine; Sahar Al Seesi; Devi Krishna Priya Karunakaran; Nikita Sturrock; Abdul Rouf Banday; Ashley M Kilcollins; Ion Măndoiu; Rahul N Kanadia; Minor splicing snRNAs are enriched in the developing mouse CNS and are crucial for survival of differentiating retinal neurons. Developmental Neurobiology 2014, 75, 895-907, 10.1002/dneu.22257.
- Marybeth Baumgartner; Anouk M Olthof; Gabriela S. Aquino; Katery C. Hyatt; Christopher Lemoine; Kyle Drake; Nikita Sturrock; Nhut Nguyen; Sahar Al Seesi; Rahul N Kanadia; et al. Minor spliceosome inactivation causes microcephaly, owing to cell cycle defects and death of self-amplifying radial glial cells. Development 2018, 145, dev166322, 10.1242/dev.166322.
- Francisco Alén; Isabel Gómez-Redondo; Patricia Rivera; Juan Suarez; Priscila Ramos-Ibeas; Eva Pericuesta; Raul Fernández-González; Serafín Perez-Cerezales; Keiko Horiuchi; Laura Orio; et al.Fernando Rodríguez De FonsecaAlfonso Gutiérrez-Adán Sex-Dimorphic Behavioral Alterations and Altered Neurogenesis in U12 Intron Splicing-Defective Zrsr1 Mutant Mice.. International Journal of Molecular Sciences 2019, 20, 3543, 10.3390/ijms20143543.
- Karen Doggett; Ben B. Williams; Sebastian Markmiller; Fan-Suo Geng; Janine Coates; Stephen Mieruszynski; Matthias Ernst; Tim Thomas; Joan Heath; Early developmental arrest and impaired gastrointestinal homeostasis in U12-dependent splicing-defective Rnpc3-deficient mice. RNA 2018, 24, 1856-1870, 10.1261/rna.068221.118.
- Yawei Gao; Xiaoyu Liu; Bin Tang; Chong Li; Zhaohui Kou; Lin Li; Wenqiang Liu; You Wu; Xiaochen Kou; Jing-Yi Li; et al.Yanhong ZhaoJiqing YinHong WangShe ChenLujian LiaoShaorong Gao Protein Expression Landscape of Mouse Embryos during Pre-implantation Development. Cell Reports 2017, 21, 3957-3969, 10.1016/j.celrep.2017.11.111.
- Lei Gao; Keliang Wu; Zhenbo Liu; Xuelong Yao; Shenli Yuan; Wenrong Tao; Lizhi Yi; Guanling Yu; Zhenzhen Hou; Dongdong Fan; et al.Yong TianJianqiao LiuZi-Jiang ChenJiang Liu Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 2018, 173, 248-259.e15, 10.1016/j.cell.2018.02.028.
- Pablo Bermejo-Álvarez; Dimitrios Rizos; D. Rath; P. Lonergan; Alfonso Gutierrez-Adan; Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proceedings of the National Academy of Sciences 2010, 107, 3394-3399, 10.1073/pnas.0913843107.
- Haihong Shen; Xuexiu Zheng; Stephan Luecke; Michael R. Green; The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes & Development 2010, 24, 2389-2394, 10.1101/gad.1974810.
- Ken-Ichiro Abe; Satoshi Funaya; Dai Tsukioka; Machika Kawamura; Yutaka Suzuki; Masataka G. Suzuki; Richard M. Schultz; F Aoki; Minor zygotic gene activation is essential for mouse preimplantation development. Proceedings of the National Academy of Sciences 2018, 115, E6780-E6788, 10.1073/pnas.1804309115.
- Toshio Hamatani; Mark G. Carter; Alexei A. Sharov; M.S. Ko; Dynamics of Global Gene Expression Changes during Mouse Preimplantation Development. Developmental Cell 2004, 6, 117-131, 10.1016/s1534-5807(03)00373-3.
- Ursula Eichenlaub-Ritter; Oocyte ageing and its cellular basis. The International Journal of Developmental Biology 2012, 56, 841-852, 10.1387/ijdb.120141ue.
- G. Flach; Martin Johnson; P.R. Braude; R.A. Taylor; V.N. Bolton; The transition from maternal to embryonic control in the 2-cell mouse embryo.. The EMBO Journal 1982, 1, 681-686, 10.1002/j.1460-2075.1982.tb01230.x.
- Geppino Falco; Sung-Lim Lee; Ilaria Stanghellini; Uwem C. Bassey; Toshio Hamatani; M.S. Ko; Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Developmental Biology 2007, 307, 539-50, 10.1016/j.ydbio.2007.05.003.
- Kei-Ichiro Ishiguro; Manuela Monti; Tomohiko Akiyama; Hiromi Kimura; Nana Chikazawa-Nohtomi; Miki Sakota; Saeko Sato; Carlo Alberto Redi; Shigeru B. H. Ko; M.S. Ko; et al. Zscan4 is expressed specifically during late meiotic prophase in both spermatogenesis and oogenesis. In Vitro Cellular & Developmental Biology - Animal 2016, 53, 167-178, 10.1007/s11626-016-0096-z.
- Sharon Schlesinger; Eran Meshorer; Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Developmental Cell 2019, 48, 135-150, 10.1016/j.devcel.2019.01.003.
- Chames Kermi; Elena Lo Furno; Domenico Maiorano; Regulation of DNA Replication in Early Embryonic Cleavages. Genes 2017, 8, 42, 10.3390/genes8010042.
- Benjamin Cieply; Juw Won Park; Angela Nakauka-Ddamba; Thomas W. Bebee; Yang Guo; Xuequn Shang; Christopher J. Lengner; Yi Xing; Russ P. Carstens; Multiphasic and Dynamic Changes in Alternative Splicing during Induction of Pluripotency Are Coordinated by Numerous RNA-Binding Proteins.. Cell Reports 2016, 15, 247-55, 10.1016/j.celrep.2016.03.025.
- Akira Hayakawa; Masao Saitoh; Keiji Miyazawa; Dual Roles for Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) in Cancer Progression. Plant Promoters and Transcription Factors 2016, 925, 33-40, 10.1007/5584_2016_50.
- Yun Huang; Jong Kyoung Kim; Dang Vinh Do; Caroline Lee; Christopher A Penfold; Jan J. Żylicz; John Marioni; Jamie A. Hackett; Azim Surani; Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. eLife 2017, 6, 3, 10.7554/eLife.22345.
- Takashi Ishiuchi; Rocio Enriquez-Gasca; Eiji Mizutani; Ana Boskovic; Céline Ziegler-Birling; Diego Rodriguez‐Terrones1; Teruhiko Wakayama; Juan M. Vaquerizas; Maria-Elena Torres-Padilla; Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nature Structural & Molecular Biology 2015, 22, 662-671, 10.1038/nsmb.3066.
- Melanie Eckersley-Maslin; Celia Alda-Catalinas; Marloes Blotenburg; Elisa Kreibich; Christel Krueger; Wolf Reik; Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program.. Genes & Development 2019, 33, 194-208, 10.1101/gad.321174.118.
- Lulzim Shkreta; Johanne Toutant; Mathieu Durand; James L Manley; Benoit Chabot; SRSF10 Connects DNA Damage to the Alternative Splicing of Transcripts Encoding Apoptosis, Cell-Cycle Control, and DNA Repair Factors.. Cell Reports 2016, 17, 1990-2003, 10.1016/j.celrep.2016.10.071.
- Zarkower, D.; Nakayama, K.; Regulation of mitosis-meiosis transition by the ubiquitin ligase beta-TrCP in male germ cells. Development 2017, 144, 4137–4147, 10.1242/dev.158485dev.158485.