Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Jason Cheng and Version 2 by Lily Guo.
Chromatin structure can either positively or negatively regulates transcription and plays an essential role in eukaryotic gene expression and cell identity.
- Chromatin Structure
1. Introduction
In eukaryotes, genomic DNA and its associated proteins, primarily histones, are organized into chromatin structural domains that affect the binding of transcription factors (TFs), recruitment of RNA polymerases (RNAPs) and the initiation and elongation of transcription [1]. The advent of new genome-wide sequencing technologies has identified chromatin-associated RNAs (ChrRNAs) [2], including nascent RNAs, coding RNAs and various regulatory non-coding RNAs (ncRNAs). RNAs can act either in cis by targeting genomic DNA or in trans through RNAP complexes and interactions with DNA-binding proteins, such as TFs and DNA/histone modifiers, to regulate chromatin structure and gene expression. There are extensive studies and reviews of the roles of ncRNAs in the regulation of DNA/histone modifications, chromatin structure, and gene expression [3][4][5][6][7][8][9][10][3,4,5,6,7,8,9,10]. In contrast, the role of RNA modifications and RNA modifying proteins (RMPs) in chromatin plasticity and transcription regulation remains largely unknown. This review will discuss recent studies, which suggest RNA modifications and RMPs function to fine-tune chromatin structure, in turn facilitating transcription activation or repression.
2. Transcription and Nascent RNA-Associated Chromatin Structure
2.1. Transcription and Nascent RNA Synthesis
Transcription is the first step of an essential multistep bioprocess and consists of three major steps: initiation, elongation and termination [11][78]. In eukaryotes, the activity between transcription initiation and elongation can be further described in five major steps: (1) chromatin remodeling; (2) formation of the preinitiation complex (PIC); (3) formation of an open complex and abortive synthesis of short (2–8) nt RNAs; (4) promoter pausing and escaping by RNAPII; and (5) pause releasing and productive elongation (Figure 1). In this section, we will describe these steps in detail.
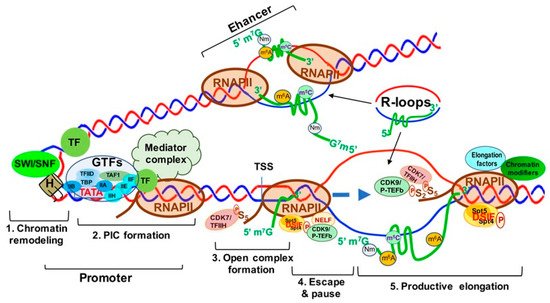
Figure 1.
Transcriptionally Active Chromatin Structure and the Five Steps of Transcription Activation.
Step 1
. Chromatin remodeling;
Step 2
. Formation of transcription pre-initiation complex (PIC);
Step 3
. Unwinding of the DNA strands to form the open complex;
Step 4
. RNA polymerase II (RNAPII) pausing and escaping from the promoter;
Step 5
. Productive transcript elongation by RNAPII. Abbreviations: SWI-SNF, switch/Sucrose non-fermentable; TF, transcription factor; H, histone; PIC, preinitiation complex; GTFs, general transcription factors; TFIIA-F, transcription factor II A–F; TAF1, TATA-box binding protein associated factor 1; TBP, TATA-box binding protein; CDK7/TFIIH, cyclin-dependent kinase 7; TSS, transcription start Site; Spt5/4, suppressor of Ty 5 and 4 (transcription elongation factors 5 and 4); DSIF, DRB sensitivity-inducing factor; CDK9/p-TEFb, cyclin-dependent kinase 9.
Chromatin remodeling begins with the interaction between sequence-specific transcription factors (TFs) and chromatin modifiers to “open” condensed chromatin (heterochromatin) [12][79] (Figure 1, step 1), allowing RNAPII/Mediator complex to bind specific DNA sequences and form a PIC at the gene promoters and enhancers [12][79] (Figure 1, step 2). The mediator of RNAPII transcription (mediator) complex is critical in transcription activation and is assembled at the PIC. Interactions between RNAPII and mediator complex are facilitated by the C-terminal domain (CTD) of the largest unit, RPB1, of RNAPII, which contains a highly conserved region with up to 52 repeats of the heptad of Y1S2P3T4S5P6S7 (YSPTSPS) largely unphosphorylated during initiation [13][80] (Figure 1, step 2). Open complex formation is initiated by the multiprotein complex TFIIH. TFIIH can not only unwind DNA to form the transcription “bubble” through its associated helicases (Figure 1, step 3), but can also phosphorylate the YSPTSPS CTD serine 5 residue through its associated CDK7 and cyclin H kinases, stimulating mediator complex and PIC general TF dissociation and allowing RNAPII escape from the promoter [14][15][81,82] (Figure 1, step 3). Two other factors, DRB sensitivity inducing factor (DSIF) and negative elongation factor (NELF) contribute to the transition from transcriptional initiation to elongation and RNAPII promoter escape [16][83]. DSIF is a heterodimer composed of Spt4 and Spt5, and NELF consists of four subunits (A, B, C/D, and E) [17][18][84,85]. Spt5 interacts with the 5′ cap of nascent RNA (pre-mRNA), and then NELF recognizes the Spt5 interface [19][86]. TFIIH interacts with DSIF and NELF, which then cooperatively induce RNAPII promoter escape and subsequent pausing (Figure 1, step 4). Finally, transcription elongation is promoted by P-TEFb [20][87], which consists of a kinase/cyclin pair of CDK9 and CCNT1 (or CCNT2). P-TEFb phosphorylates DSIF and RNAPII CTD serine 2, inducing NELF release from RNAPII [21][88] and allowing RNAPII interaction with other elongation factors and chromatin modifying complexes to ensure productive transcription elongation, respectively [22][23][24][89,90,91] (Figure 1, step 5).
2.2. R-Loops as the Regulators of Transcription and Chromatin
2.2.1. Mechanisms of R-Loop Formation
R-loops are three-strand nucleic acid structures consisting of an RNA-DNA duplex and an unpaired DNA strand generated during transcription upon nascent RNA “threadback” invasion into the DNA duplex to displace the non-template strand [25][92] (Figure 1). Different from the RNA: DNA hybrids occurring inside the active sites of RNAP during replication and transcription, R-loops span a much longer range (~100–200 base pairs) and form outside of RNAP during transcription [26][93]. R-loops are found ubiquitously from bacteria to mammals and frequently at gene promoters and enhancers, as well as terminators. Functionally, R-loops can positively and negatively regulate transcription and affect the differentiation and lineage plasticity of pluripotent progenitors/stem cells [27][28][29][94,95,96].
2.2.2. Distribution and Function of R-Loops
R-loops prevent DNA methylation by DNMT3B1 and are found to be clustered at the CpG islands (CGIs) of gene promoters and enhancers in the human genome [30][97]. In transcription activation, R-loops are found to promote recruitment of epigenetic modifiers and RNAPII to CGIs [31][98]. As shown in Figure 1, R-loops at gene promoters and enhancers are associated with active chromatin markers, such as H3 lysine 27 acetylation (H3K27ac) and H3 lysine 4 trimethylation (H3K4me3), which suggests the involvement of R-loops in enhancer–promoter looping and high-order chromatin architecture [28][32][95,99].
Antisense RNA transcripts promote the formation of R-loops, creating a local chromatin environment which facilitates the recruitment of transcription factors and chromatin modifiers to promote gene transcription [33][34][100,101]. For example, the LncRNA TARID is reportedly involved in TCF21 transcriptional activation by generating an R-loop at the CpG-rich promoter of TCF21, enabling GADD45A binding to the R-loop and recruitment of the DNA demethylase TET1 [35][102]. On the other hand, it is also well-documented that R-loops can trigger chromatin condensation and heterochromatin formation to repress gene expression [36][103]. Co-transcriptional R-loops have been shown to impede the movement of RNAP in vitro and exert a negative impact on transcription elongation [37][104]. R-loops were reported to regulate the chromatin structure of specific genes involved in control of mouse ESC differentiation by differentially interacting with the active Tip60-p400 histone acetyltransferase complex and repressive PRC2 H3K27me3 complex [28][95]. Mechanistically, higher Tip60-p400 and lower PRC2 levels are associated with promoter-proximal R-loops and active transcription to promote ESC differentiation, while decreased Tip60-p400 and increased PRC2 disrupts R-loop formation and impairs ESC differentiation [28][95].
In addition to gene promoters and enhancers, R-loops are highly enriched in retrotransposons (i.e., LINE-1), ribosomal RNA (rRNA), tRNA loci and mitochondria, and are crucial for regulating genome stability, stress response and mitochondrial function [38][39][40][105,106,107]. R-loops can cause hypermutation, hyperrecombination, chromosomal rearrangements or chromosome loss, leading to genome instability and chromosome fragility [41][108]. Genome-wide mapping of R-loops demonstrates an enrichment at the 3′ end of genes with GC skew patterns in mammalian cells, and R-loop accumulation at the 3′ end of genes inhibits transcription [42][109]. R-loops were found to be associated with a subset of developmental regulator genes, which are targeted by Polycomb complexes PCR1 and PCR2 [43][110]. Removal of the R-loop causes decreased PRC1 and PRC2 binding and increased RNAPII recruitment, resulting in activation of these PRC1/2 targeting genes [43][110]. Arginine methyltransferases PRMT1/5 and CARM1 interact with tudor domain-containing protein 3 (TDRD3) to facilitate recruitment of topoisomerase IIIB (TOP3B) to prevent such R-loop accumulation [44][111], and release RNAPII for successive transcription by methylating the arginine residues in histone tails, as well as the RNAPII CTD [45][112].
It is increasingly clear that the formation, distribution and removal of R-loops is tightly regulated during DNA transcription and replication. R-loops are involved in regulation of all stages of gene expression, from transcription initiation to termination, and can act as transcriptional activators or repressors at specific gene loci in different cell types and differentiation states. Regulatory R-loops are important for transcription elongation and termination, as well as telomere stability and DNA repair in normal physiological conditions. In contrast, dysregulated R-loops are associated with transcription elongation defects, DNA damage and genome instability in pathophysiological conditions, such as cancer [25][38][92,105].
2.3. MYC, 7SK snRNP and BRD4 Transcription and Chromatin Structure Regulation
2.3.1. MYC-Mediated RNA 5’ Capping, Transcription Elongation and Active Chromatin
MYC is well-known to activate Myc-dependent transcription, but its underlying mechanisms remain incompletely understood [46][47][113,114]. As shown in Figure 26, MYC is involved in m7G 5′ capping of pre-mRNAs by binding to the E-box in the promoter of active genes, which recruits the capping enzyme complex composed of RNA guanylyltransferase and 5’-phosphatase (RNGTT) and CMTR1 [47][114]. Upon binding to the E-box, MYC also recruits CDK7/TFIIH and CDK9/P-TEFb to gene promoters, which phosphorylate RNAPII CTD serine 5 and 2 residues, respectively [48][115].
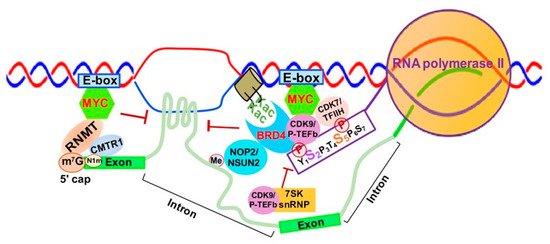
Figure 26.
MYC-mediated RNA 5′ Capping, Transcription Elongation and Chromatin Structure. Abbreviations: E-box- enhancer box; MYC- MYC proto-oncogene BHLH transcription factor; RNMT, RNA methyltransferase; CMTR1, cap methyltransferase 1; K-ac, acetylated lysine; CDK9/P-TEFb, cyclin-dependent Kinase 9; CDK7/TFIIH, Cyclin-dependent Kinase 7; BRD4, Bromodomain Containing 4; 7SK snRNP, 7SK small nuclear ribonucleoprotein.
Finally, MYC appears to be implicated in regulation of R-loop formation as well. Recent studies have shown that MYC can suppress R-loop formation in promoter-proximal regions through recruitment of BRCA1, which stabilizes the DCP2 de-capping enzyme complex and enables RNAPII escape from promoter pausing, leading to increased transcription elongation and gene expression [49][116] (Figure 26).
2.3.2. SK snRNP Complex as a Negative Regulator of Transcription Elongation
The 7SK small nuclear ribonucleoprotein (7SK snRNP) complex is composed of the core 7SK snRNPs and other proteins, including AFF1/4, HEXIM1/HEXIM2, LARP7, MePCE and hnRNPs [50][117]. This complex functions as a major negative regulator of CDK9/P-TEFb by inhibiting CDK9/P-TEFb-mediated RNAPII escape and transcription elongation [51][52][118,119]. 7SK snRNP interacts with HEXIMI that directly binds CDK9/P-TEFb, inactivating its kinase activity and inhibiting transcription elongation [53][120] (Figure 26).
2.3.3. BRD4-Mediated Transcription Elongation and Active Chromatin
BRD4 is a member of the Bromodomains and Extraterminal (BET) family and contains two bromodomains that recognize acetylated lysine residues of histone tails [54][55][121,122]. BRD4 not only promotes the establishment of chromatin, favoring transcriptional activation, but also directly competes with release of CDK9/P-TEFb sequestrated by the 7SK snRNP complex, which promotes CTD-S2 phosphorylation and activates RNAPII-dependent transcription elongation [56][123] (Figure 26). BRD4 was initially reported to interact with CDK9/P-TEFb through its bromodomain, resulting in CDK9/P-TEFb-dependent phosphorylation of RNAPII CTD serine 2 (CTD-S2) and transcription activation in vivo [57][58][124,125]. However, later studies demonstrated that BRD4 functions as an atypical kinase by directly binding and phosphorylating CTD-S2 both in vitro and in vivo under conditions where other CTD kinases are inactive [59][126]. Our study demonstrated that RNA cytosine methyltransferase NOP2/NSUN1 and NSUN2 interact with BRD4 and the elongating form of RNAP (CTD-S2P) to form a transcriptionally active chromatin structure in leukemia cells [60][127] (Figure 26). BRD4 was also found to prevent R-loop accumulation and to protect against transcription-replication collision [61][128]. Loss of BRD4 in cancer cells leads to replication stress, DNA damage and eventually apoptotic cell death [61][128].
3. Fine-Tuning Chromatin and Transcription through RNA Modifications and RNA-Modifying Proteins
3.1. m
6
A and RMPs Modulate Interactions between Non-Coding RNAs, Transcription Factors and Chromatin Modifiers
The METTL3/14 methyltransferases, YTH-domain-containing proteins (YTHDs)/HNRNPs, and FTO/ALKB family proteins function as m6A writers, readers and erasers, respectively, to regulate the level and distribution of m6A in a tissue-/differentiation-specific manner [62][63][64][65][66][31,34,38,46,216] (Figure 2A). Recent studies have shown widespread m6A in human tissues, which is significantly enriched in long intergenic non-coding RNA (lincRNA) and CpG-rich gene promoters, suggesting that m6A is positively correlated with gene expression homeostasis and has broad involvement in human development and disease [67][217].
Increasing evidence suggests that m6A and its associated RMPs, more specifically m6A writers and readers, modulate chromatin structure and transcriptional activation through interactions with TFs and chromatin modifying proteins/complexes [68][69][218,219]. METTL14 was reported to recognize the active histone mark H3K36me3, resulting in METTL3-METTL14-WTAP m6A methyltransferase complex recruitment to the vicinity of H3K36me3 peaks, promoting m6A and positively regulating transcription elongation [70][220] (Figure 38A).
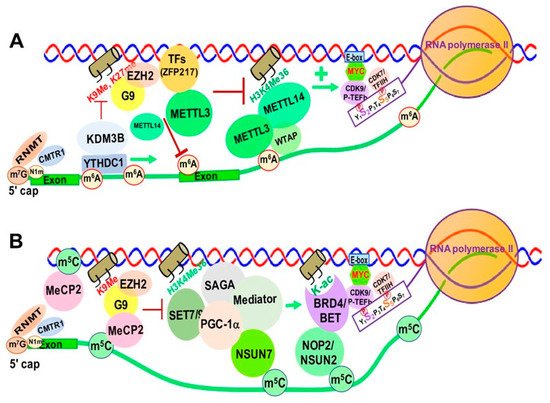

Figure 38. RNA Modification/RMP-mediated, Transcription-associated Chromatin Structural Changes in Mammalian Cells. (A). m6A and its RMPs-mediated chromatin structural changes. (B). m5C and its RMPs-mediated chromatin structural changes. Abbreviations: m6A, N6-methyladenine; RNMT, RNA Methyltransferase; CMTR1, cap methyltransferase 1; KDM3B, lysine demethylase 3B; YTHDC1, YTH Domain Containing 1; EZH2, enhancer of Zeste 2 Polycomb repressive Complex 2 Subunit; METTL3/14, methyltransferase Like 3/14; K9me, lysine 9 methyl; K27me, lysine 27 methyl; TFs, Transcription Factors; ZFP217, zinc finger Protein 217; H3K4me36, histone 3 lysine 36 tri-methylation; E-box, enhancer box; CDK9/P-TEFb, cyclin-dependent kinase 9; CDK7/TFIIH, cyclin-dependent kinase 7. m5C, 5-methylcytosine; MeCP2, methyl-CpG binding protein 2; SET7/9, SET Domain containing 7/9 histone lysine methyltransferase; SAGA, Spt-Ada-Gcn5-acetyltransferase; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1-α; NSUN7, NOP2/Sun RNA methyltransferase member 7; H3K4me36, histone 3 lysine 36 tri-methylation; NOP2/NSUN1, NOP2 nucleolar protein/NOP2/Sun RNA methyltransferase 1; K-ac, lysine acetylation; BRD5/BET, bromodomain containing 4; E-box, enhancer box; CDK9/pTEFb, cyclin-dependent kinase 9; CDK7/TFIIH, cyclin-dependent Kinase 7; MYC, MYC proto-oncogene BHLH transcription factor.
Zinc finger protein 217 (ZFP217), a chromatin-associated oncogenic TF important for embryonic stem cell (ESC) differentiation [71][221], is associated with repressive chromatin marked by histone 3 lysine 9 trimethylation (H3K9me3) and histone 3 lysine 27 trimethylation (H3K27me3) through interactions with G9a and EZH2 methyltransferases [8]. ZFP217 also negatively affects m6A deposition at its own target gene mRNAs by sequestering METTL3 in an inactive status, in which METTL3 is dissociated from the METTL14/WTAP co-factor complex [71][72][221,222] (Figure 38A).
YTHDC1 can de-repress (activate) gene expression by physically interacting with and recruiting KDM3B demethylase to hyper-m6A in nascent RNAs, leading to demethylation of the repressive transcriptional histone modification H3K9me2 [73][223]. YTHDC1 can also facilitate the decay of a subset of these m6A-modified RNAs, especially LINE-1, through nuclear exosome targeting-mediated nuclear degradation, while METTL3 deposits m6A modifications on chromosome-associated regulatory RNAs (carRNAs), including promoter-associated RNAs, enhancer RNAs and repeat RNAs [74][224].
m6A was reported to promote translation of the SETD1A/B components, which increases the transcriptional activation histone mark H3K4me3 and recruitment of the erythroid-specific zinc finger transcription factor KLF1 to its target gene promoters [75][225]. Inhibition of the METTL3/METTL14/WTAP m6A methyltransferase complex blocks erythropoiesis in human bone marrow hematopoietic stem/progenitor cells (HSPCs). The METTL3-METTL14-WTAP complex also functionally interacts with SMAD2/3 proteins, the key signal transducers and transcriptional modulators of the TGFβ pathway. SMAD2/3 promotes binding of the m6A methyltransferase complex to a subset of transcripts involved in early cell fate decisions [76][226]. In addition, a recent study showed that RBM15, a subunit of the m6A methyltransferase complex, mediates the degradation of the pre-BAF155 mRNA to affect the chromatin remodeling function of the BAF (SWI/SNF-like) complex [77][227].
3.2. The Impact of m
5
C and Its RMPs on Chromatin and Transcription
In eukaryotes, RNA m5C is catalyzed by RCMTs, including NOL1/NOP2/SUN domain (NSUN) family enzymes and DNMT2 (Figure 2B). NSUN2 is responsible for catalyzing the majority of m5C in mRNAs and non-coding RNAs [78][79][80][57,228,229]. Our study demonstrated a marked increase in NOP2/NSUN1 and NSUN2-mediated m5C in drug (5-azacitidine) resistant myeloid leukemia cells [60][127]. NSUN7 is reported to promote transcription of genes labeled with enriched m5C eRNAs in mouse hepatoma cells and primary hepatocytes for adaptive metabolic responses [76][226]. Mechanistically, the methyltransferase SET7/9 methylates histone 3 K4 residues [77][227] and PGC-1α K779, a transcriptional co-activator important for regulating adapted metabolic responses [81][230]. Methylated PGC-1α interacts with Spt-Ada-Gcn5-acetyltransferase (SAGA) and mediator complexes, as well as NSUN7, at the m5C-marked enhancer RNAs of PGC-1α target genes to promote recruitment of transcription machinery and reinforce transcription of those genes (Figure 38).
Heterozygous mutations in the X-linked gene encoding MeCP2 cause the neurological disorder Rett syndrome. MeCP2 interacts with more than 40 binding partners, including transcriptional regulators, chromatin modifiers and RNA- splicing factors [82][231]. MeCP2 chromatin binding is controlled by m5C, and recent studies show enrichment of MeCP2 on long non-coding RNAs (lncRNAs), such as retinal noncoding RNA3 (RNCR3) [83][232], muscle-specific long non-coding RNA (ChRO1) [84][233], and major Satellite Forward RNAs [85][234]. This MeCP2 enrichment on lncRNAs promotes deposition of H3K9me3 and H4K20me3 and heterochromatin formation. Additionally, m5C is present in a wide variety of RNA species, including cytoplasmic and mitochondrial ribosomal RNAs (rRNAs) and tRNAs, as well as mRNAs, eRNAs and other types of non-coding RNAs [86][55]. There are widespread and dynamic changes in RNA m5C in response to stress and stimuli, which suggests an important role of m5C and its RMPs in epigenetic gene regulation [86][55]. However, the detailed mechanisms underlying m5C and its RMP-mediated chromatin remodeling and gene regulation are yet to be elucidated.
3.3. RNA Modifications and RMPs in R-Loop Formation and Transcription Regulation
R-loops can be formed both in cis by a nascent RNA folding back to its DNA template at the same transcription locus and in trans by ncRNAs at distant enhancers (Figure 5). R-loops are prevalent in highly transcribed genes, and occur most frequently at conserved genetic hotspots, such as unmethylated CpG island promoters, G-rich terminators and other regulatory chromosomal loci [31][37][98,104]. R-loops play an important role in organizing chromatin structure and maintaining genome stability [32][99], and RNA modifications and their associated RMPs can modulate the interactions between chromatin-associated RNAs and chromatin modifiers, as well as transcription machinery, to regulate chromatin structure and transcription in a position and context-dependent manner [87][235].
3.3.1. m6A and Its RMPs Promote R-Loop Formation at Transcription Termination Sites
A recent study showed that R-loops accumulate in m6A-rich transcription termination sites (TTSs) [88][236]. Depletion of m6A methyltransferase complex components dramatically reduces R-loop accumulation in m6A marked genes around TTSs, leading to disruption of proper transcription termination [88][236]. In contrast, depletion of the nuclear m6A reader protein YTHDC1 does not affect R-loop levels [88][236].
3.3.2. m6A and RMPs Associated R-Loop Formation on DNA Damage-Associated RNAs
ATM serine/threonine kinase (ATM) is recruited to DNA double-strand break (DSB) sites to phosphorylate several key proteins, leading to cell cycle arrest, DNA repair or apoptosis [89][237]. Specifically, in response to DSBs, ATM phosphorylates METTL3 serine 43, and the phosphorylated METTL3 is then recruited to DNA-damage sites to catalyze m6A in the DNA damage-associated RNAs. This increased m6A deposition leads to YTHDC1 recruitment and R-loop accumulation, promoting homologous recombination-mediated DNA repair [90][238]. In human pluripotent stem cells, m6A-containing R-loops are reported to accumulate during G2/M and are depleted at G0/G1 phases of the cell cycle [91][239]. Knockout of the m6A reader YTHDF2 leads to increased R-loop levels, cell growth retardation, and accumulation of the DNA double-strand break marker γH2AX in mammalian cells, which suggests that YTHDF2 negatively regulates m6A and R-loop accumulation [91][239]. Taken together, these results suggest that m6A and its RMPs regulate accumulation of R-loops and genomic stability in a position and context-dependent manner [29][92][93][96,240,241].
3.3.3. RNA Cytosine Methylations and RMPs in R-Loop Formation
R-loops have been found to be required for transcription-coupled homologous recombination, as they recruit Cockayne Syndrome Protein B (CSB) and RAD52 proteins, but not the canonical HR proteins BRCA1 and BRCA2, to sites of reactive oxygen species (ROS)-induced DNA damage [94][242]. The formation of R-loops at break sites is dependent upon Drosha, an RNase III enzyme in the microRNA (miRNA) biogenesis apparatus [95][243]. DNMT2 is predominantly located in the cytoplasm and mitochondria, where it contributes to m5C in tRNAs, consequently affecting protein translation [96][97][98][244,245,246]. Little is known about the role of DNMT2 in regulating chromatin structure and R-loop formation. A recent report showed that DNMT2 (Figure 3B) is recruited to DNA damage sites, where it deposits m5C onto the DNA damage-associated mRNAs and contributes to R-loop formation. The m5C and DNMT2-mediated R-loops facilitate the homologous recombination (HR) and DNA repair of DSBs [99][247].
Loss of TRDMT1 in cancer cells confers sensitivity to PARP inhibitors in vitro and in vivo, suggesting that transcription-coupled RNA modifications may serve as DNA damage markers to regulate DNA repair [99][247]. METTL8 was previously reported to contribute m3C in mRNA, while METTL2 and METTL6 contribute m3C in tRNAs [100][248]. Recently, METTL8 was found to form a large SUMOylated nuclear RNA-binding protein complex that is associated with R-loops, and knockout of METTL8 resulted in reduction of m3C and R-loops in the nucleolus [101][249]. The nucleolar-enriched SUMOylated METTL8 complex regulates R-loop formation and promotes tumorigenesis [101][249]. Retention of R-loops at their transcription sites of nascent RNAs is an important and intrinsic part of chromatin structure in human cells, and R-loop dysregulation is associated with DNA damage, transcription elongation defects, hyper-recombination and genome instability [38][102][105,250].
3.4. RNA Editing in Chromatin Remodeling
3.4.1. A-To-I Editing and ADAR-Mediated Heterochromatin and Gene Silencing
A-to-I editing is one of the most abundant RNA modifications in mammalian cells and is catalyzed by ADAR family proteins ADAR1 and ADAR2 (Figure 3A). A-to-I editing occurs most frequently in non-coding RNAs within repetitive elements in the genome, mainly Alu repeats [103][104][105][251,252,253], and has a critical role in immune regulation [106][107][69,254], and alternative splicing [108][67]. Vigilin is the largest protein in the KH domain-containing RNA-binding protein family [109][255]. Vigilin binds to promiscuously A-to-I-edited RNAs, predominantly in inverted Alu repeats, and is involved in the formation of heterochromatin [110][256], which is marked with the repressive chromatin marker H3K9me3 by histone H3-specific methyltransferase SUV39H1 [111][112][257,258]. Mechanistically, vigilin binds to A-to-I-edited RNAs in a complex that contains ADAR1, as well as RNA helicase A and Ku86/70 [113][259]. The C-terminal domain of human vigilin interacts with SUV39H1 and recruits it to the A-to-I-edited RNAs, leading to formation of heterochromatin and gene silencing [113][259]. This suggests a mechanistic link between DNA-repair machinery and the A-to-I RNA editing complex. Interestingly, the levels of global A-to-I editing and m6A in mammalian cells were shown to be negatively correlated, and depletion of m6A modification increases the association of m6A-depleted transcripts with ADAR enzymes, resulting in upregulated A-to-I editing on the same m6A-depleted transcripts [114][115][72,260].
3.4.2. C-To-U Editing and Associated RMPs in Chromatin Remodeling and Gene Regulation
C-to-U editing is carried out by the APOBECs family proteins (Figure 3B). APOBECs function to restrict retroviruses and retrotransposons [116][261], and have been identified as a prominent source of mutations in cancers [117][262]. APOBEC3B (A3B) was shown to regulate expression of the estrogen receptor α (ERα)-target gene by causing C-to-U deamination at ER binding regions [118][263]. Further, transient cytidine deamination by A3B promotes chromatin modification and remodeling at regulatory regions of ER target genes that increase ER expression levels, and elevated A3B expression is associated with poor patient survival in ER+ breast cancer [118][263]. However, more studies are needed to understand the role of C-to-U editing and the APOBECs in regulation of chromatin structure and gene expression.
