You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Hai Liu.
Jasmonic acid (JA), a lipid-derived molecule, plays an essential function in controlling many different plant developmental and stress responses.
- phytohormone
- jasmonic acid
- JA signaling
1. Introduction
During growth and development, plants are constantly battling against a challenging environment. These adverse or unfavorable environmental conditions are often categorized as: (i) abiotic stresses, such as ultraviolet (UV) radiation, flood, drought, heat, cold, heavy metal toxicity and nutrient deficiency, and (ii) biotic stresses, such as pathogen infection and animal herbivory [1]. Within plant cells, various signal transduction pathways work collaboratively to convey and integrate stress stimuli, and ultimately orchestrate processes of plant growth, development and defense responses [2,3,4][2][3][4]. Phytohormones are among the most important signaling molecules that are involved in the signaling network that regulates these processes [5,6,7,8,9,10,11,12][5][6][7][8][9][10][11][12].
Jasmonic acid (JA) and its metabolic derivatives, such as jasmonic acid isoleucine (JA-Ile) and methyl jasmonate (MeJA), collectively known as jasmonates (JAs), are a class of lipid-derived, natural and widely distributed phytohormones in higher plants. JAs have been studied for decades as key signaling compounds involved in many aspects of plant development and stress responses [9,13,14,15,16,17,18][9][13][14][15][16][17][18]. Upon stress stimuli, such as wounding, herbivory or necrotrophic pathogen infection, plant cells trigger a rapid increase of JAs, which lead to the activation of defense responses and reproduction, as well as the inhibition of growth for plant fitness [19,20,21,22,23][19][20][21][22][23]. Moreover, through the crosstalk network, JAs often work in concert with other phytohormones, such as abscisic acid (ABA), auxin, cytokinin (CK), ethylene (ET), gibberellic acid (GA) and salicylic acid (SA), to balance between growth- and defense-related processes, thereby conferring plants acclimation to the changing environments [10,11,24][10][11][24].
2. JA Signaling
2.1. JA Perception and Signal Transduction
The generally accepted “relief of repression” model for JA perception is built upon decades of research beginning with the identification of the core co-receptor complex for JA-Ile, that is composed of the F-box protein CORONATINE INSENSITIVE 1 (COI1) containing SKP1-CULLIN1-F-box-type (SCF) E3 ubiquitin ligase complex SCFCOI1, JASMONATE ZIM DOMAIN (JAZ) proteins and inositol pentakisphosphate (InsP5) [34,35,36,57,58,59,60][25][26][27][28][29][30][31]. Under normal conditions, where little or no nuclear JA-Ile is present, certain TFs, such as MYC2 (a basic helix-loop-helix (bHLH) family TF and key activator of JA responses), are repressed by a series of JASMONATE ZIM DOMAIN (JAZ) proteins through direct interaction. MYC2 binds to the G-box motif at the promoter regions of the JA-responsive genes and activates their expression [61][32] (Figure 21). Most JAZ family members have been shown to interact with MYC2. When binding to MYC2, the JAZ protein recruits the TOPLESS (TPL) and TPL-related (TPR) co-repressors directly or through the adaptor protein NOVEL INTERACTOR OF JAZ (NINJA) to repress the transcriptional activity of MYC2 (Figure 21). The transcriptional repression function of the TPL co-repressors involves the further recruitment of the chromatin modifying HISTONE DEACETYLASE (HDA) complex that “switches off” the targeted region by chromatin condensation [62,63,64][33][34][35]. Members of HDAs such as HDA6 and HDA19 have been shown to participate in JA responses [65,66][36][37].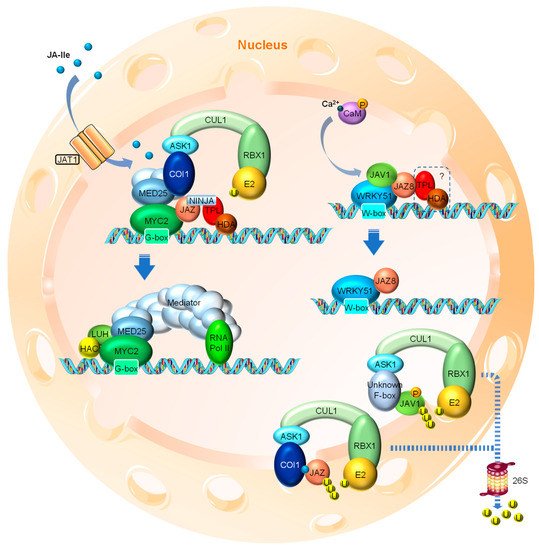
Figure 21. Simplified model of JA signaling in Arabidopsis. When nuclear bioactive JA levels are low, the master transcription factors, such as MYC2, are repressed through the interaction with JAZ proteins that recruit other co-repressors, such as NINJA, TPL and HDA, to form a repressor complex at the promoter regions of JA-responsive genes. In addition, MED25 also physically interacts with MYC2 while bound with COI1, the F-box subunit of the SCFCOI1 E3 ubiquitin ligase complex. In the case of the JA biosynthesis gene AOS, its transcription is repressed by the JJW complex composed of JAV1, JAZ8 and WRKY51. In both cases, the expression of JA-responsive genes is restrained. It is unclear whether the JJW complex also recruits co-repressors, such as TPL and HDA. When a certain developmental or environmental cue triggers the import of bioactive JA (e.g., JA-Ile) into the cell nucleus presumably through the action of JAT1, elevated levels of JA-Ile cause the formation of COI1-JA-JAZ co-receptor complex. The interaction between COI1 and JAZ leads to the dissociation of JAZ and MYC2, as well as the dissociation of COI1 and MED25. As a result, JAZ is degraded via the 26S proteasome and the enhanced interaction between MED25 and MYC2 ultimately leads to MED25-mediated transcriptional activation of the target genes. In the case of JJW-regulated AOS, stress-induced fast Ca2+ influx leads to the CaM-mediated phosphorylation of JAV1. JAV1 phosphorylation causes the disintegration of the JJW complex and AOS transcriptional activation. Phosphorylated JAV1 is subjected to E3 ubiquitin ligase-mediated ubiquitination and 26S proteasomal degradation, although the F-box protein responsible for the specific recognition of JAV1 remains to be identified. JAZ, JASMONATE ZIM DOMAIN; NINJA, NOVEL INTERACTOR OF JAZ; TPL, TOPLESS; HDA, HISTONE DEACETYLASE; MED25, MEDIATOR25; COI1, CORONATINE INSENSITIVE1; ASK1, ARABIDOPSIS SKP1-RELATED1; CUL1, CULLIN1; SCF, SKP1-CULLIN1-F-box; HAC1, HISTONE ACETYLTRANSFERASE1; LUH, LEUNIG_HOMOLOG; JAV1, JASMONATE ASSOCIATED VQ DOMAIN PROTEIN1; AOS, ALLENE OXIDE SYNTHASE; CaM, Calmodulin. RBX1 is a RING finger protein that recruits the E2 ubiquitin-conjugating enzyme to the C-terminus of CUL1.
2.2. JA-Regulated Transcription Factors
In addition to MYC2 serving as the main transcriptional regulator of JA-induced gene activation, other members of the MYC TF family as well as members of other TF families have also been shown to be directly involved in controlling JA-regulated gene expression (Figure 32). MYC3 and MYC4 are also targets of JAZ repressors (e.g., JAZ3 and JAZ5) and act additively with MYC2 to activate JA response in the vegetative tissue, especially the JA-dependent defense response against wounding and herbivory [87,88][60][61]. MYC2, MYC3, MYC4 and MYC5 interact with at least two R2R3-MYB TFs, MYB21 and MYB24, to form a MYC-MYB transcription complex (Figure 32). Both MYC and MYB are repressed by JAZ suppressor and are activated by JA to cooperatively regulate stamen development in Arabidopsis [89,90,91,92][62][63][64][65]. In rice, data have also shown that the JA-responsive R2R3-type MYB TFs, JAMYB and its homolog, are transcription activators directly regulated by JA [93][66]. JAMYB binds to the AG-motif-like motif in the promoter region of Argonaute18 (AGO18) gene, which encodes a core RNA silencing component that promotes AGO1-mediated antiviral RNAi [94][67]. The transactivation activity of JAMYB is normally repressed by JAZ6. The JA accumulation elicited by rice stripe virus coat protein triggers the ubiquitination and proteasomal degradation of JAZ6, relieving the repression of JAMYB to activate the expression of AGO18. Elevated accumulation of AGO18 ultimately leads to enhanced antiviral defense in rice [93,94][66][67]. It is reasonable to hypothesize that certain rice MYC homologs also interact with JAMYB.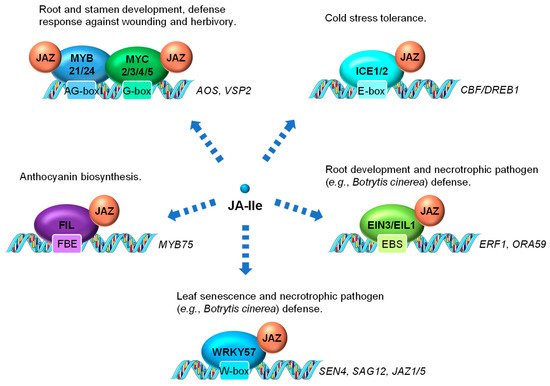
Figure 32. Examples of JA-regulated processes control through the interaction between JAZ and transcription factors (TFs). The MYC TFs, several R2R3-MYB family members (including MYB21 and MYB24), and other TFs (e.g., ICE1, EIN3, EIL1, and FIL) are direct targets of JAZ repressors. These TFs are activated by JA-mediated JAZ degradation and positively regulate JA responses. WRKY57 is also a direct target of JAZ but acts as a negative regulator of JA regulated leaf senescence and defense against necrotrophs. ICE1, INDUCER OF CBF EXPRESSION1; EIN3, ETHYLENE INSENSITIVE3; EIL1, EIN3-LIKE1; FIL, FILAMENTOUS FLOWER; EBS, EIN3 binding site; FBE, FIL DNA binding element.
3.3. Negative Feedbacks and Termination of JA Signal
2.3. Negative Feedbacks and Termination of JA Signal
Since JA is a stress signal that generally leads to growth inhibition, proper desensitization and termination of the JA signal is undoubtedly as important as its activation for overall plant growth and fitness. In fact, the JA signal is elaborately controlled at multiple levels to ensure that each response only lasts for an appropriate period at an appropriate amplitude [30,80,103][76][53][77]. Cytosolic JA-Ile dynamics are shaped by at least two JA-inducible catabolic pathways in Arabidopsis. The first pathway is the direct oxidation of JA-Ile by members of the cytochrome P450 subfamily 94 (CYP94) enzymes, CYP94B1, CYP94B3 and CYP94C1, which turn JA-Ile into bio-inactive 12OH-JA-Ile and 12COOH-JA-Ile [104,105] (Figure 1)[78][79]. The second pathway is the deconjugation of JA-Ile mediated by two amidohydrolases, IAR3 and ILL6, which hydrolyze both JA-Ile and 12OH-JA-Ile [106] (Figure 1)[80]. Both pathways have been shown to contribute additively for the turnover of JA-Ile but act differently for JA responses and tolerance to related stress conditions [103,107][77][81]. In crop plants (e.g., rice and corn), JA catabolism has also been proven to be crucial to both the development, such as sexual determination [108,109][82][83], and stress tolerances, such as salt and cold [110,111][84][85]. JA-Ile stimulates rapid activation of JAZ gene expression while most JAZ genes in Arabidopsis can produce truncated JAZ splice variants that can still bind to the MYC proteins but have little capability of forming complexes with JA-Ile and COI1 for proteasomal degradation. Overexpression of certain JAZ splice variants, such as JAZ10, result in dominant repression of JA responses [112,113][86][87]. Crystal structure reveals that the JAZ10 splice variant tightly binds to MYC3 and blocks the interaction between MYC3 and MED25, which is crucial for the transcriptional activation of MYC3 target genes [114][88]. These findings indicate that the alternative splicing of JAZ genes serve as a general feedback mechanism to desensitize JA signaling. Intriguingly, JA also induces the recruitment of two splicing factors, PRE-mRNA-PROCESSING PROTEIN 39a (PRP39a) and PRP40a to JAZ loci by MED25. PRP39a and PRP40a, in turn, facilitate the full splicing of JAZ transcripts to produce the full-length JAZ proteins, thus preventing the excessive desensitization of JA responses caused by the overaccumulation of JAZ splice variants [115][89]. These data suggest that the JA-induced negative feedback mechanism by the alternative splicing of JAZ genes is under exquisite modulation. Several bHLH family subgroup IIId members (e.g., the JASMONATE-ASSOCIATED MYC2-LIKE proteins (JAMs) in Arabidopsis and the MYC2-TARGETED BHLHs (MTBs) in tomato) have been identified as negative regulators of JA responses [116,117,118,119][90][91][92][93]. In Arabidopsis, JAM1/bHLH17, JAM2/bHLH13 and JAM3/bHLH3 interact with JAZs and function as transcriptional repressors by competing with MYCs for G-box binding [116,117,118][90][91][92]. Likewise, the tomato MTB1, MTB2 and MTB3 are activated by MYC2 but act in turn to negatively regulate JA responses by competing with MYC2 for the target promoter binding site (i.e., the G-box motif), impeding the formation of the MYC2-MED25 complex [119][93]. In addition to the activation of JAMs or MTBs, JA also stabilizes BTB/POZ-MATH3 (BPM3), one of the BPM proteins that function as adaptors of Cullin3-based E3 ubiquitin ligases [120][94]. Several BPMs are found to directly target MYC2, MYC3 and MYC4 for polyubiquitination and degradation. Thus, the stabilities of JA-activated MYCs are negatively controlled by BPMs, especially BPM3, whose stability is greatly enhanced by JA [120][94]. Taken together, these groundbreaking discoveries suggest that plant cells orchestrate a complex and autoregulatory negative feedback circuit to desensitize and terminate JA signals at multiple layers.References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324.
- Sierla, M.; Waszczak, C.; Vahisalu, T.; Kangasjarvi, J. Reactive Oxygen Species in the Regulation of Stomatal Movements. Plant Physiol. 2016, 171, 1569–1580.
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431.
- Fichman, Y.; Mittler, R. Rapid systemic signaling during abiotic and biotic stresses: Is the ROS wave master of all trades? Plant J. 2020, 102, 887–896.
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119.
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176.
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496.
- Saeed, W.; Naseem, S.; Ali, Z. Strigolactones Biosynthesis and Their Role in Abiotic Stress Resilience in Plants: A Critical Review. Front. Plant Sci. 2017, 8, 1487.
- Wang, J.; Wu, D.; Wang, Y.; Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 2019, 70, 3391–3400.
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349.
- Jang, G.; Yoon, Y.; Choi, Y.D. Crosstalk with Jasmonic Acid Integrates Multiple Responses in Plant Development. Int. J. Mol. Sci. 2020, 21, 305.
- Zhou, Y.; Xiong, Q.; Yin, C.C.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene Biosynthesis, Signaling, and Crosstalk with Other Hormones in Rice. Small Methods 2020, 4.
- Katsir, L.; Chung, H.S.; Koo, A.J.; Howe, G.A. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008, 11, 428–435.
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058.
- Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 2017, 37, 42–47.
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321.
- Acosta, I.F.; Przybyl, M. Jasmonate Signaling during Arabidopsis Stamen Maturation. Plant Cell Physiol. 2019, 60, 2648–2659.
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479.
- Glauser, G.; Grata, E.; Dubugnon, L.; Rudaz, S.; Farmer, E.E.; Wolfender, J.L. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 2008, 283, 16400–16407.
- Koo, A.J.; Gao, X.; Jones, A.D.; Howe, G.A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009, 59, 974–986.
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575.
- Wasternack, C.; Strnad, M. Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds. Int. J. Mol. Sci. 2018, 19, 2539.
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621.
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359.
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671.
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665.
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405.
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094.
- Yan, Y.; Stolz, S.; Chetelat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483.
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236.
- Mosblech, A.; Thurow, C.; Gatz, C.; Feussner, I.; Heilmann, I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011, 65, 949–957.
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245.
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703.
- Yamamuro, C.; Zhu, J.K.; Yang, Z. Epigenetic Modifications and Plant Hormone Action. Mol. Plant 2016, 9, 57–70.
- Jiang, J.; Ding, A.B.; Liu, F.; Zhong, X. Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 2020, 71, 5179–5190.
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.W.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234.
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544.
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350.
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100.
- Shyu, C.; Figueroa, P.; Depew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 2012, 24, 536–550.
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679.
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791.
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca(2+)/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149.e7.
- Li, C.J.; Shi, L.; Wang, Y.N.; Li, W.; Chen, B.Q.; Zhu, L.; Fu, Y. Arabidopsis ECAP Is a New Adaptor Protein that Connects JAZ Repressors with the TPR2 Co-repressor to Suppress Jasmonate-Responsive Anthocyanin Accumulation. Mol. Plant 2020, 13, 246–265.
- Cevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555.
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916.
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, E8930–E8939.
- Poss, Z.C.; Ebmeier, C.C.; Taatjes, D.J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 575–608.
- Yang, Y.; Li, L.; Qu, L.J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2016, 58, 106–118.
- Zhai, Q.; Li, C. The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424.
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273.
- You, Y.; Zhai, Q.; An, C.; Li, C. LEUNIG_HOMOLOG Mediates MYC2-Dependent Transcriptional Activation in Cooperation with the Coactivators HAC1 and MED25. Plant Cell 2019, 31, 2187–2205.
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86.
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.C.; Cai, H.Y.; Qin, Y.; Mullis, A.; Lin, Z.G.; Zhang, L.S. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2017, 36, 311–335.
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13.
- Hu, P.; Zhou, W.; Cheng, Z.; Fan, M.; Wang, L.; Xie, D. JAV1 controls jasmonate-regulated plant defense. Mol. Cell 2013, 50, 504–515.
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Simura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356.
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245.
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782.
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715.
- Zhang, C.; Lei, Y.; Lu, C.; Wang, L.; Wu, J. MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 2020, 62, 1159–1175.
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013.
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633.
- Huang, H.; Gao, H.; Liu, B.; Qi, T.; Tong, J.; Xiao, L.; Xie, D.; Song, S. Arabidopsis MYB24 Regulates Jasmonate-Mediated Stamen Development. Front. Plant Sci. 2017, 8, 1525.
- Song, S.S.; Huang, H.; Wang, J.J.; Liu, B.; Qi, T.C.; Xie, D.X. MYC5 is Involved in Jasmonate-Regulated Plant Growth, Leaf Senescence and Defense Responses. Plant Cell Physiol. 2017, 58, 1752–1763.
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate Signaling Enhances RNA Silencing and Antiviral Defense in Rice. Cell Host Microbe 2020, 28, 89–103 e108.
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733.
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015, 56, 7–15.
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924.
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369.
- Goossens, J.; Fernandez-Calvo, P.; Schweizer, F.; Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689.
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725.
- Boter, M.; Golz, J.F.; Gimenez-Ibanez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate. Plant Cell 2015, 27, 3160–3174.
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814.
- Wen, J.; Li, Y.; Qi, T.; Gao, H.; Liu, B.; Zhang, M.; Huang, H.; Song, S. The C-terminal domains of Arabidopsis GL3/EGL3/TT8 interact with JAZ proteins and mediate dimeric interactions. Plant Signal. Behav. 2018, 13, e1422460.
- Gupta, A.; Bhardwaj, M.; Tran, L.P. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. Int. J. Mol. Sci. 2020, 21, 7482.
- Heitz, T.; Smirnova, E.; Marquis, V.; Poirier, L. Metabolic Control within the Jasmonate Biochemical Pathway. Plant Cell Physiol. 2019, 60, 2621–2628.
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738.
- Aubert, Y.; Widemann, E.; Miesch, L.; Pinot, F.; Heitz, T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 2015, 66, 3879–3892.
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J. Biol. Chem. 2013, 288, 31701–31714.
- Marquis, V.; Smirnova, E.; Poirier, L.; Zumsteg, J.; Schweizer, F.; Reymond, P.; Heitz, T. Stress- and pathway-specific impacts of impaired jasmonoyl-isoleucine (JA-Ile) catabolism on defense signalling and biotic stress resistance. Plant Cell Environ. 2020, 43, 1558–1570.
- Lunde, C.; Kimberlin, A.; Leiboff, S.; Koo, A.J.; Hake, S. Tasselseed5 overexpresses a wound-inducible enzyme, ZmCYP94B1, that affects jasmonate catabolism, sex determination, and plant architecture in maize. Commun. Biol. 2019, 2, 114.
- Wang, F.; Yuan, Z.J.; Zhao, Z.W.; Li, C.X.; Zhang, X.; Liang, H.F.; Liu, Y.W.; Xu, Q.; Liu, H.T. Tasselseed5 encodes a cytochrome C oxidase that functions in sex determination by affecting jasmonate catabolism in maize. J. Integr. Plant Biol. 2020, 62, 247–255.
- Kurotani, K.; Hayashi, K.; Hatanaka, S.; Toda, Y.; Ogawa, D.; Ichikawa, H.; Ishimaru, Y.; Tashita, R.; Suzuki, T.; Ueda, M.; et al. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015, 56, 779–789.
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.Y.; Yu, Y.; Li, L.; Peng, C.; Fan, T.; et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501.
- Chung, H.S.; Cooke, T.F.; Depew, C.L.; Patel, L.C.; Ogawa, N.; Kobayashi, Y.; Howe, G.A. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010, 63, 613–622.
- Moreno, J.E.; Shyu, C.; Campos, M.L.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017.
- Zhang, F.; Ke, J.; Zhang, L.; Chen, R.; Sugimoto, K.; Howe, G.A.; Xu, H.E.; Zhou, M.; He, S.Y.; Melcher, K. Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 1720–1725.
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2020, 32, 429–448.
- Sasaki-Sekimoto, Y.; Jikumaru, Y.; Obayashi, T.; Saito, H.; Masuda, S.; Kamiya, Y.; Ohta, H.; Shirasu, K. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 2013, 163, 291–304.
- Song, S.; Qi, T.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.; Guo, H.; Xie, D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013, 9, e1003653.
- Fonseca, S.; Fernandez-Calvo, P.; Fernandez, G.M.; Diez-Diaz, M.; Gimenez-Ibanez, S.; Lopez-Vidriero, I.; Godoy, M.; Fernandez-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS ONE 2014, 9, e86182.
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127.
- Chico, J.M.; Lechner, E.; Fernandez-Barbero, G.; Canibano, E.; Garcia-Casado, G.; Franco-Zorrilla, J.M.; Hammann, P.; Zamarreno, A.M.; Garcia-Mina, J.M.; Rubio, V.; et al. CUL3(BPM) E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses. Proc. Natl. Acad. Sci. USA 2020, 117, 6205–6215.
More
