The signaling pathways of brassinosteroids (BRs), a unique plant steroid hormone, are critically involved in a diverse range of plant growth and developmental processes as well as many important agronomic traits.
- brassinosteroids
- BR signaling components
- rice
- Arabidopsis
- plant development
- crop yield
1. Introduction
Brassinosteroids (BR) regulate a diverse spectrum of processes in plant growth and development as well as internal modulation in response to environmental fluctuation. Since they are essential in plant adaptation and seed plant evolution, the biosynthesis and signaling pathways have been identified in the model plant Arabidopsis, providing a comprehensive understanding of how BR synthesis is controlled and how the signaling pathways are coordinated during the plant’s life cycle [1][2][3][4][5][6][1,2,3,4,5,6]. Moreover, BR controls a number of important agronomic traits including plant height, grain size, tillering, leaf angle, and environmental adaptations, exploiting the potential of the BR response controlling genes as an interesting target by which to enhance crop performance [7][8][7,8].
2. BR Signaling Pathway in Dicot and Monocot Model System
Brassinosteroids were originally isolated from rape (Brassica napus) pollen in 1979 [9]. After identification of BR as a plant-specific steroid hormone, extensive genetic and biochemical studies revealed its signal transduction mechanism in Arabidopsis. In detail, the initiation of BR signaling is tightly mediated by a receptor-like kinase, BRASSINOSTEROID INSENSITIVE 1 (BRI1), and co-receptor kinase, BRI1-ASSOCIATED KINASE 1 (BAK1), at the plasma membrane [1][4][1,4]. BR perception through these receptor complexes triggers the dissociation of a negative regulator, BRI1 KINASE INHIBITOR 1 (BKI1), and confers a transphosphorylation of BRI1 and BAK1, leading to the activation of BRI1 SUPPRESSOR 1 (BSU1) and consequent inactivation of BRASSINOSTEROID INSENSITIVE 2 (BIN2) kinase, a representative of the plant GLYCOGEN SYNTHASE KINASE 3 (GSK3) [1][2][3][4][10][1,2,3,4,10]. The BIN2 strictly regulates the phosphorylation status of plant-specific transcription factors, BRASSINAZOLE-RESISTANT 1 (BZR1) and BR-INSENSITIVE-EMS-SUPPRESSOR 1/BRASSINAZOLE-RESISTANT 2 (BES1/BZR2), which play critical roles in BR perception downstream events via specific binding to the cis-element in the promoter region of large target genes [5][6][11][5,6,11]. In the absence of BR, BIN2 is activated by auto-phosphorylation and directly phosphorylates BZR1 and BES1, leading to cytosolic accumulation through 14-3-3 binding and degradation by 26S-proteasome [12][13][14][12,13,14].
In the case of the BR signaling pathway in rice, the physiological role of BR and potential BR signaling components have been investigated since the 1980s. Among the diverse primary BR responses (plant height, grain size, lamina bending, grain filling, stress resistance) in rice, the degree of the lamina joint angle serves as a model system by which to determine BR response and sensitivity and contributes to the extensive identification of BR biosynthesis and/or signaling components in rice [15][16][15,16]. As a result of intense screening using lamina joint angle and plant height, the primary BR signaling components, the orthologue of Arabidopsis BRI1 in rice (OsBRI1), was first characterized as a functional BR receptor through forward genetic screening as a typical BR-defective phenotype such as erect leaf and dwarfism. Among the various alleles of the OsBRI1 gene, the d61-4 mutant, which carries a null mutation of the BR receptor, shows severely reduced second-node elongation and lamina joint angle but has a mild effect on the plant’s fertility. The second component of BR signaling, OsBAK1, also characterized as an orthologue of Arabidopsis BAK1, functions as a BR co-receptor with BRI1 as evidenced by its loss-of-function mutation, and it displays an erect leaf phenotype and BR insensitivity [17]. In addition, ectopic expression of OsBRI1 or OsBAK1 complements the BR-defective phenotype of d61, supporting the notion that OsBRI1 and OsBAK1 function as BR co-receptors in rice [18]. After the perception of BR in the plasma membrane in rice, OsBRI1/OsBAK1 transduces the phosphorylation cascade and consequently inactivates GLYCOGEN SYNTHASE KINASE (GSK), OsGSK1, and OsGSK2 [19][20][19,20]. These Arabidopsis BIN2 counterparts in rice mainly suppress the BR response through direct phosphorylation of rice BR transcription factors. The loss-of-function mutation of OsGSK1/2 results in increased BR sensitivity to lamina inclination and plant height, highlighting the conserved functional role in rice BR signaling [21]. In addition, the rice genome contains four OsBZR1s encoding the rice counterpart of Arabidopsis BZR1, a key transcription factor regulating BR-responsive gene expression. Similar to the phosphorylation-dependent regulation of BZR1 in Arabidopsis, OsBZR1 physically interacts with and is phosphorylated by OsGSKs, which induces the cytoplasmic retention of OsBZR1 via the 14-3-3 protein in rice [22]. OsBZR1 is also controlled by the OsPUB24 ubiquitin ligase through targeted degradation to balance BR signaling homeostasis [23]. Consistent with its positive role in BR signaling, the silencing of OsBZR1 in rice results in semi-dwarfism and decreased lamina joint angle, suggesting that OsBZR1 acts as a downstream BR signaling component in rice.
Interestingly, rice BR signaling involves a diverse range of transcriptional regulators which modulate the primary BR-responsive genes. These include DWARF AND LOW-TILLERING (DLT), LEAF AND TILLER ANGLE INCREASED CONTROLLER (LIC), GRAIN LENGTH 2/GROWTH REGULATING FACTOR 4 (GL2/OsGRF4), OVATE FAMILY PROTEIN (OFP), REDUCED LEAF ANGLE 1/SMALL ORGAN SIZE 1 (RLA1/SMOS1), and U-TYPE CYCLIN (CYC-U), which fine-tune the BR response spatiotemporally via direct and/or indirect interaction with canonical BR signaling components (OsBRI1, OsBAK1, OsGSKs, OsBZR1) in rice (Figure 1). The DLT is a transcriptional regulator that belongs to the GRAS gene family and is phosphorylated by OsGSK2 in a BR-dependent manner. In the presence of BR, the accumulation of the hypo-phosphorylated form of DLT induces several BR responses, such as cell elongation and lamina inclination. Interestingly, BR negatively regulates DLT expression through OsBZR1 activation, but DLT induces OsBZR1 to maintain BR signaling homeostasis [20][24][20,24]. The loss-of-function dlt displays the typical BR-deficient phenotype, namely, semi-dwarfism, erect leaves, and reduced tiller number, but overexpression of DLT induces increased BR sensitivity and a physiological BR response. The DLT is also involved in the BR biosynthetic pathway in the context of feedback regulation in controlling the expression of key BR biosynthesis enzymes D2 and DWARF, suggesting that DLT is another positive transcriptional regulator of BR signaling in rice [7][25][7,25].
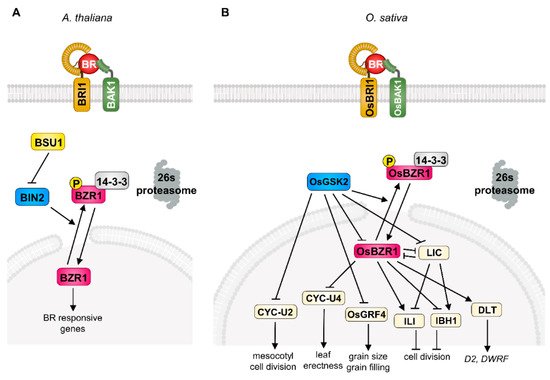
Figure 1.
Arabidopsis
Arabidopsis
A
B
Arabidopsis
A
B
The CCCH-type zinc-finger protein, LIC, is another important transcriptional regulator in rice BR signaling. The silencing and ectopic expression of LIC resulted in elevated and compromised physiological BR responses in rice, respectively. In addition to genetic evidence, the biochemical study of OsGSK2 with LIC also supports its role in the BR signal transduction pathway. The LIC directly interacts with OsGSK2 and is phosphorylated and subsequently accumulated in the cytosol. As a consequence, BR deprivation leads to the loss of the transcriptional activity of LIC and results in enhanced BR sensitivity as a feedback mechanism. Importantly, OsBZR1 directly binds to the promoter of LIC and represses its expression in a BR-dependent manner, indicating the antagonistic relationship between LIC and OsBZR1 in rice BR signaling. Moreover, this relationship seems to be manifested in differential responses to low or high BR concentrations; a low physiological concentration of BR preferentially activates BR signaling via OsBZR1, but a high BR concentration mainly functions to repress the BR response in the LIC-dependent signaling pathway, which potentially aids in rice adaptation via precise BR responses in development and stress resistance [26][27][26,27].
The OsGRF4 is another key player controlling BR-responsive gene expression in rice. OsGRF4 was originally characterized as a positive regulator of grain size and filling in rice through grain-size-associated quantitative trait loci (QTL) analysis. Interestingly, OsGSK2 directly interacts with and represses OsGRF4 transcriptional activity under the control of BR perception and miR396 also targets the OsGRF family, indicating that OsGRF4 is specifically involved in grain-related BR responses and is an integrator of diverse hormonal crosstalk in rice [28][29][30][31][28,29,30,31].
The OFP has been recently identified as a transcription factor that interacts with the DLT protein. In the absence of BR, the OsGSK2 kinase attenuates OFP’s transcriptional activity, whereas BR induces the accumulation of OFP protein in the nucleus and positively regulates the BR response in rice in a DLT-dependent manner [32]. Interestingly, ectopic expression of OFP stimulates gibberellin (GA) inactivation enzyme expression, suggesting that OFP serves as the locus of crosstalk in the context of the BR-induced GA inactivation process in rice [8].
A recent study identified a rice-specific interaction partner of OsBZR1. The APETALA2 DNA-binding transcription factor RLA/SMOS1 forms a complex with OsBZR1 for full activation of BR signal transduction. OsGSK2 also phosphorylates and inactivates, but auxin stimulates RLA/SMOS1 expression, indicating possible crosstalk between BR and auxin signal transduction [33][34][33,34].
The CYC-U2 protein, U-type cyclin, is also an important OsGSK2 kinase substrate promoting cell division in the mesocotyl of rice. In the presence of BR, inactivation of OsGSK2 leads to mesocotyl cell division [35]. The RELATED TO ABI3/VP1-LIKE 1 (RAVL1) is another BR signaling component that is required for full activation of BR signaling. The expression level of OsBRI1 is regulated by RAVL1, which directly controls the BR synthesis genes D2, D11, and BRASSINOSTEROID DEFICIENT 1 (BRD1) through binding to the E-box on its target gene’s promoter region [7]. The loss-of-function of RAVL1 is manifested by semi-dwarfism, delayed germination of seedlings, and a dark green phenotype, supporting the notion of its positive effect on rice BR responses [36].
In general, the identification of canonical BR signaling pathway components in rice has greatly improved the understanding of the BR’s functional mechanism for enhancing crop performance. It is noteworthy that OsBZR1 directly/indirectly interacts with a number of TF complexes, such as DLT, LIC, OsGRF4OFP, RLA1/SMOS1, and CYC-U4;1, and these interaction partners are also largely controlled by OsGSKs in an OsBRI1-dependent manner. In the context of different BR levels in specific tissues and stages, differential assembly combination of OsBZR1-interacting transcriptional regulators will generate a diverse series of BR responses or specific BR responses in different tissues/stages during the rice life cycle.
