Liquid Biopsy (LB) is a novel method for cancer diagnosis performed by analyzing and sampling of non-solid biological tissues, obtained primarily from blood, but also from other body fluids such as urine, saliva and cerebrospinal fluid.
- pancreatic ductal adenocarcinoma
- liquid biopsy
- ctDNA
- exosomes
- CTCs
- miRNAs
1. Introduction
Tumors and their metastases release biomarkers, mainly CTCs, cell free nucleic acids (cfDNA and cfRNAs), extracellular vesicles such as exosomes, and tumor educated platelets (TEPs), that can distantly reflect the disease (Figure 1Figure 2). Therefore, liquid biopsies (LBs) represent a minimally invasive technique and allow diagnosis, real-time monitoring of cancer evolution and molecular follow-up of patients [1][2]. Also, LBs give us a better picture of the tumor heterogeneity than a tissue biopsy which only captures a specific area, since the whole tumor mass releases material into the blood [3].
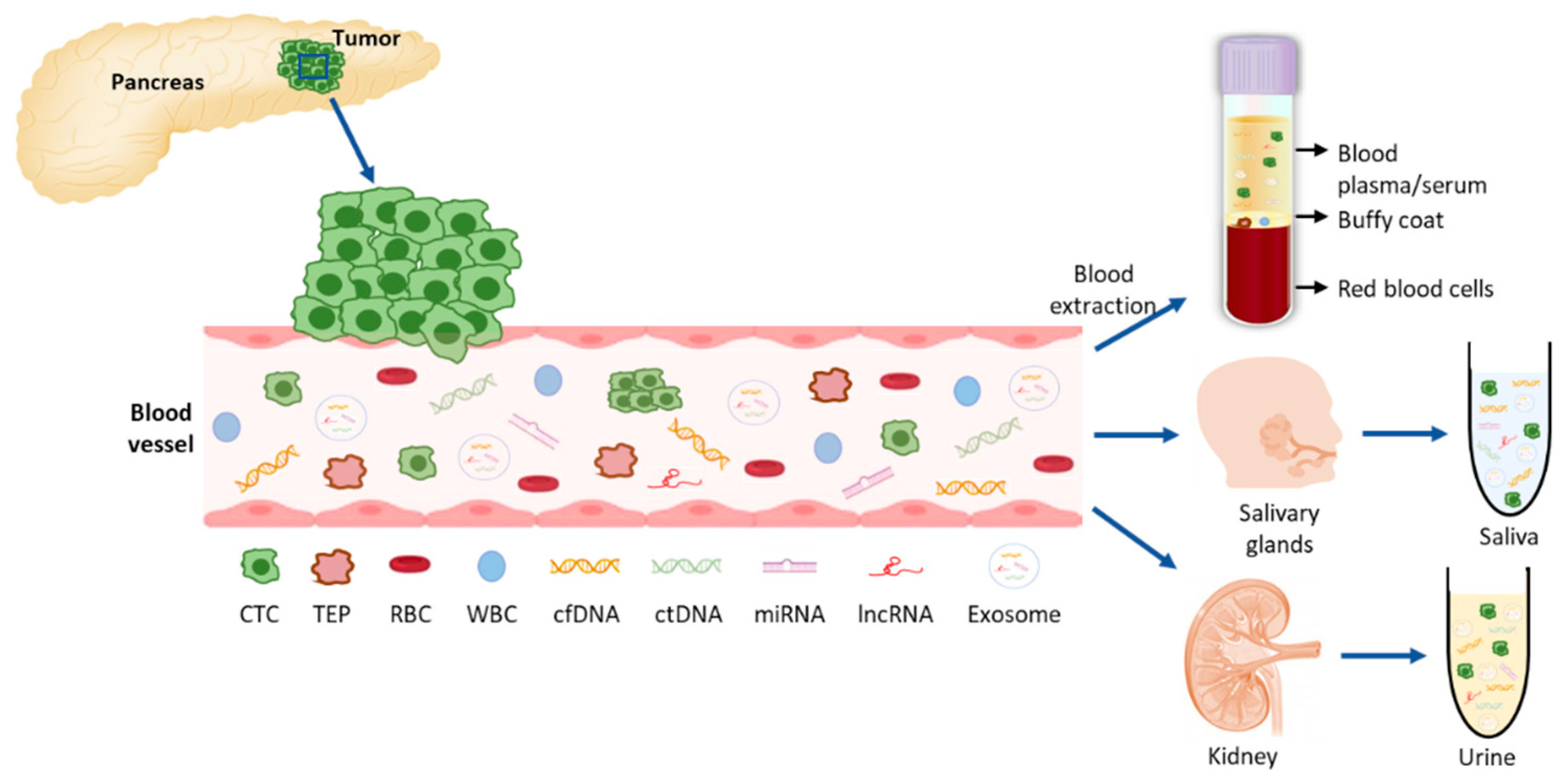
Figure 12.
Recent technological and molecular advances have increased our ability to detect and analyze LB components. In the following lines we will briefly introduce the different methods currently available for blood-based LBs (Table 1).
Table 1. Methods for isolation and analysis of liquid biopsy components in pancreatic cancer. Overview of the advantages and disadvantages of the described methods.
| LB Component | Technique | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| cfDNA | qPCR | Fast & Inexpensive High specificity |
Lower sensitivity (0.1%) Detects only point mutations |
[4][5][6][7][8][9][10] |
| dPCR* (ddPCR, BEAMING) | High sensitivity (0.01%) & specificity | Detects only point mutations Expensive |
||
| NGS | High DNA input permits high throughput analysis and screen for unknown variants (WGS &WES) Can identify structural variants and copy number variations |
Variable sensitivity (0.1% aprox.) Expensive |
||
| Exosomes | Density-based isolation* (centrifugation) | Inexpensive Independent of marker expression |
Time consuming High volume sample required Can damage exosomes Contaminated sample |
[11][1][2][12][13][14][15][16] |
| Size-based isolation | Fast & Inexpensive Independent of marker expression |
Contaminated sample | ||
| Affinity-based isolation | High purity and specificity | Low sample yield | ||
| Commercial kits | Fast & Simple | Expensive | ||
| CTCs | Immunoaffinity enrichment* | Positive enrichment: - Very specific - High capture efficiency & purity Negative enrichment: - Label-free CTCs obtained |
Only one subpopulation captured Lower purity |
[11][4][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31] |
| Physical methods (size & density) | Represent tumor heterogeneity Fast & Simple Less expensive Label-free CTCs obtained |
Must be followed with immuno-labelling techniques to distinguish CTCs | ||
| cfRNAs | RT-qPCR | Fast & Inexpensive High specificity |
Low sensitivity in samples with low abundance cfRNA | [32] |
| ddPCR* | Higher sensitivity & accuracy Lower sample volume required More reproducible than qPCR |
Tedious assay optimization |
2. Liquid Biopsy in Other Body Fluids for the Early Detection of Pancreatic Ductal Adenocarcinoma (PDAC)
Molecular analyses for the early detection of PDAC via LB are also being developed in other body fluids such as pancreatic juice, saliva and urine. The collection of pancreatic juice from the duodenal lumen is less invasive than other tissue biopsy collection methods, but it is still a relative invasive and difficult technique that needs to be performed by specialized personnel. Nevertheless, a number of molecular studies have been performed for the diagnosis of PDAC using pancreatic juice, mainly for the detection of KRAS mutations [33]. A meta-analysis of 16 studies that analyzed the diagnostic value of KRAS mutations revealed that the sensitivity and specificity levels for the diagnosis of PDAC were 0.59 and 0.87, respectively [34], and another meta-analysis of 39 studies assessing the diagnostic value of the four major altered genes in PDAC (KRAS, CDKN2A, TP53 and SMAD4), telomerase activity, and combination assay revealed that the most reliable marker was telomerase activity with a sensitivity and specificity of 0.82 and 0.96, respectively [35]. However, these results should be analyzed with caution since they cannot distinguish early PDAC from intraductal papillary mucinous neoplasm (IPMN), or pancreas with low-grade PanIN, since alterations of KRAS and telomerase activity are also found in these lesions [33].
Saliva is a very convenient fluid for LB determinations since it can be easily and noninvasively obtained from patients, and it has been reported that it contains almost the same molecules as serum because of the high blood flow in salivary glands [5]. Molecular studies have been performed in saliva for the diagnosis of PDAC. Exosomes have been found in saliva in preclinical models and have been reported to discriminate PDAC, hence, they might be potential biomarkers for detecting PDAC [36]. A salivary transcriptomic analysis has identified a four-messenger RNA panel (MBD3L2, KRAS, ACRV1 and DPM1) that discriminates patients with PDAC from healthy controls with 0.9 sensitivity and 0.95 specificity [37]. MiRNAs miR-3679-5p and miR-940 have been reported to be down- and up-regulated in PDAC compared to healthy controls and benign lesions. The combined analysis of these miRNAs showed 0.7 sensitivity and 0.7 specificity in PDAC vs. noncancer [38]. The same group evaluated the expression of salivary lncRNAs and identified up-regulated levels of HOTAIR and PV1T in PDAC patients in comparison to healthy controls, with a combined sensitivity and specificity of 0.78 and 0.91 respectively. These values raised to 0.82 sensitivity and 0.95 specificity when differentiating PDAC from benign tumors [39].
Urine can be viewed as an ultrafiltrate of plasma and therefore may contain biomarkers that could assist with PDAC diagnosis [5]. Urine LB has the main advantages of allowing a completely non-invasive sampling and high volume collection, and has a lower proteome content than blood to avoid contamination of possible biomarkers [40]. Because of this, many metabolomic [41][42][43][44] and proteomic [45][46][47][48] studies have been conducted in order to identify possible biomarkers that can aid in the early identification of PDAC. With this purpose, Debernardi et al. have reported a urinary biomarker panel comprising LYVE1, REG1B, and TFF1 and PancRISK score that can discriminate patients with early stages of PDAC from control individuals and patients with benign hepatobiliary diseases [49].
Regarding cell free nucleic acids, detection of KRAS mutations in urine from PDAC patients has also been reported, and the detection rate and sensitivity are comparable to plasma LB [50]. Urinary miRNA biomarkers have also been analyzed and significant over-expression of miRNAs in PDAC Stage I versus healthy individuals (miR-143, miR-223, miR-30e) and Stage I versus Stages II-IV PDAC (miR-204, miR-143, miR-223) have been described [51]. A recent study also showed that the miR-3940-5p/miR-8069 ratio in urine exosomes is elevated in PDAC patients, suggesting that it may be a potential diagnostic tool for PDAC, especially in combination with CA19.9 [52].
3. Conclusions
Although there were many expectations set on the use of LB for early diagnosis, the truth is that the relatively low sensitivity and specificity of current techniques does not allow its use for these purposes. In addition, the available studies suggest that patients with PDAC in whom ctDNA is detected at the time of diagnosis have a poor prognosis and have a high chance of relapse after surgery, so it is advisable to develop more sensitive techniques that allow diagnosing tumors in earlier stages and provide patients with a better prognosis. It is possible that, with the development of ultrasensitive techniques, the joint use of different biomarkers and epigenetic marks, the sensitivity of LB will increase without losing specificity, and LB could be applied in PDAC screening.
The different LB methods have been shown to be a reliable biomarker in relation to the prognosis of patients with PDAC for both PFS and OS. Furthermore, its variations throughout treatment predict response or resistance to treatment several weeks in advance, so it could be used to guide treatment based on the evolution of the biomarker. In addition, the study of the characteristics of CTCs, ctDNA, exoDNA and miRNA can help us to better characterize the tumor and to identify potential therapeutic targets that facilitate the selection of treatment.
However, it is necessary to standardize and validate the methodology to be used in the different LB modalities. In addition, the usefulness of other LB modalities should be explored, such as lncRNAs or TEPs. On the other hand, the usefulness of less invasive sources of ctDNA such as urine or saliva needs to be further investigated.
Finally, it should be remembered that in order to apply LB to clinical practice, it is necessary to reduce costs, standardize protocols, and have data generated in the context of large-scale prospective clinical trials that confirm that the information provided contributes significantly to improving therapeutic results in PDAC patients.
References
- Buscail, E.; Maulat, C.; Muscari, F.; Chiche, L.; Cordelier, P.; Dabernat, S.; Alix-Panabieres, C.; Buscail, L. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers 2019, 11, 852.
- Junqueira-Neto, S.; Batista, I.A.; Costa, J.L.; Melo, S.A. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019, 63, 479–488.
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34.
- Lee, J.S.; Park, S.S.; Lee, Y.K.; Norton, J.A.; Jeffrey, S.S. Liquid biopsy in pancreatic ductal adenocarcinoma: Current status of circulating tumor cells and circulating tumor DNA. Mol. Oncol. 2019, 13, 1623–1650.
- Zhou, B.; Xu, J.W.; Cheng, Y.G.; Gao, J.Y.; Hu, S.Y.; Wang, L.; Zhan, H.X. Early detection of pancreatic cancer: Where are we now and where are we going? Int. J. Cancer 2017, 141, 231–241.
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535.
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136–168.
- Rothe, F.; Laes, J.F.; Lambrechts, D.; Smeets, D.; Vincent, D.; Maetens, M.; Fumagalli, D.; Michiels, S.; Drisis, S.; Moerman, C.; et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 1959–1965.
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554.
- Stahlberg, A.; Krzyzanowski, P.M.; Egyud, M.; Filges, S.; Stein, L.; Godfrey, T.E. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat. Protoc. 2017, 12, 664–682.
- Kamyabi, N.; Bernard, V.; Maitra, A. Liquid biopsies in pancreatic cancer. Expert Rev. Anticancer Ther. 2019, 19, 869–878.
- Coumans, F.A.; van der Pol, E.; Boing, A.N.; Hajji, N.; Sturk, G.; van Leeuwen, T.G.; Nieuwland, R. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J. Extracell. Vesicles 2014, 3, 25922.
- Woo, H.K.; Sunkara, V.; Park, J.; Kim, T.H.; Han, J.R.; Kim, C.J.; Choi, H.I.; Kim, Y.K.; Cho, Y.K. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. ACS Nano 2017, 11, 1360–1370.
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723.
- Grant, R.; Ansa-Addo, E.; Stratton, D.; Antwi-Baffour, S.; Jorfi, S.; Kholia, S.; Krige, L.; Lange, S.; Inal, J. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J. Immunol. Methods 2011, 371, 143–151.
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 2011, 728, 235–246.
- Banko, P.; Lee, S.Y.; Nagygyorgy, V.; Zrinyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48.
- Miltenyi, S.; Muller, W.; Weichel, W.; Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 1990, 11, 231–238.
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975.
- Lu, N.N.; Xie, M.; Wang, J.; Lv, S.W.; Yi, J.S.; Dong, W.G.; Huang, W.H. Biotin-triggered decomposable immunomagnetic beads for capture and release of circulating tumor cells. Acs Appl. Mater. Interfaces 2015, 7, 8817–8826.
- Tong, X.; Xiong, Y.; Zborowski, M.; Farag, S.S.; Chalmers, J.J. A novel high throughput immunomagnetic cell sorting system for potential clinical scale depletion of T cells for allogeneic stem cell transplantation. Exp. Hematol. 2007, 35, 1613–1622.
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239.
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397.
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Gottert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641.
- Gleghorn, J.P.; Pratt, E.D.; Denning, D.; Liu, H.; Bander, N.H.; Tagawa, S.T.; Nanus, D.M.; Giannakakou, P.A.; Kirby, B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip 2010, 10, 27–29.
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStream(), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 24133.
- Di Trapani, M.; Manaresi, N.; Medoro, G. DEPArray system: An automatic image-based sorter for isolation of pure circulating tumor cells. Cytom. Part A J. Int. Soc. Anal. Cytol. 2018, 93, 1260–1266.
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.I.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014, 9, 694–710.
- Lu, Y.T.; Zhao, L.; Shen, Q.; Garcia, M.A.; Wu, D.; Hou, S.; Song, M.; Xu, X.; Ouyang, W.H.; Ouyang, W.W.; et al. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods 2013, 64, 144–152.
- Campton, D.E.; Ramirez, A.B.; Nordberg, J.J.; Drovetto, N.; Clein, A.C.; Varshavskaya, P.; Friemel, B.H.; Quarre, S.; Breman, A.; Dorschner, M.; et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer 2015, 15, 360.
- Rosenberg, R.; Gertler, R.; Friederichs, J.; Fuehrer, K.; Dahm, M.; Phelps, R.; Thorban, S.; Nekarda, H.; Siewert, J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49, 150–158.
- Drula, R.; Ott, L.F.; Berindan-Neagoe, I.; Pantel, K.; Calin, G.A. MicroRNAs from Liquid Biopsy Derived Extracellular Vesicles: Recent Advances in Detection and Characterization Methods. Cancers 2020, 12, 2009.
- Satoh, K. Molecular Approaches Using Body Fluid for the Early Detection of Pancreatic Cancer. Diagnostics 2021, 11, 375.
- Yang, J.; Li, S.; Li, J.; Wang, F.; Chen, K.; Zheng, Y.; Wang, J.; Lu, W.; Zhou, Y.; Yin, Q.; et al. A meta-analysis of the diagnostic value of detecting K-ras mutation in pancreatic juice as a molecular marker for pancreatic cancer. Pancreatol. Off. J. Int. Assoc. Pancreatol. 2016, 16, 605–614.
- Hata, T.; Ishida, M.; Motoi, F.; Yamaguchi, T.; Naitoh, T.; Katayose, Y.; Egawa, S.; Unno, M. Telomerase activity in pancreatic juice differentiates pancreatic cancer from chronic pancreatitis: A meta-analysis. Pancreatol. Off. J. Int. Assoc. Pancreatol. 2016, 16, 372–381.
- Lau, C.; Kim, Y.; Chia, D.; Spielmann, N.; Eibl, G.; Elashoff, D.; Wei, F.; Lin, Y.L.; Moro, A.; Grogan, T.; et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 2013, 288, 26888–26897.
- Zhang, L.; Farrell, J.J.; Zhou, H.; Elashoff, D.; Akin, D.; Park, N.H.; Chia, D.; Wong, D.T. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010, 138, 949–957.e7.
- Xie, Z.; Yin, X.; Gong, B.; Nie, W.; Wu, B.; Zhang, X.; Huang, J.; Zhang, P.; Zhou, Z.; Li, Z. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev. Res. 2015, 8, 165–173.
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419.
- Adachi, J.; Kumar, C.; Zhang, Y.; Olsen, J.V.; Mann, M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006, 7, R80.
- Napoli, C.; Sperandio, N.; Lawlor, R.T.; Scarpa, A.; Molinari, H.; Assfalg, M. Urine metabolic signature of pancreatic ductal adenocarcinoma by (1)h nuclear magnetic resonance: Identification, mapping, and evolution. J. Proteome Res. 2012, 11, 1274–1283.
- Davis, V.W.; Schiller, D.E.; Eurich, D.; Bathe, O.F.; Sawyer, M.B. Pancreatic ductal adenocarcinoma is associated with a distinct urinary metabolomic signature. Ann. Surg. Oncol. 2013, 20 (Suppl. S3), S415–S423.
- Lusczek, E.R.; Paulo, J.A.; Saltzman, J.R.; Kadiyala, V.; Banks, P.A.; Beilman, G.; Conwell, D.L. Urinary 1H-NMR metabolomics can distinguish pancreatitis patients from healthy controls. JOP J. Pancreas 2013, 14, 161–170.
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; Gonzalez Maldonado, S.; Pilarsky, C.; Heidecke, C.D.; Schatz, P.; et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2018, 67, 128–137.
- Roy, R.; Zurakowski, D.; Wischhusen, J.; Frauenhoffer, C.; Hooshmand, S.; Kulke, M.; Moses, M.A. Urinary TIMP-1 and MMP-2 levels detect the presence of pancreatic malignancies. Br. J. Cancer 2014, 111, 1772–1779.
- Hogendorf, P.; Durczynski, A.; Skulimowski, A.; Kumor, A.; Poznanska, G.; Strzelczyk, J. Neutrophil Gelatinase-Associated Lipocalin (NGAL) concentration in urine is superior to CA19-9 and Ca 125 in differentiation of pancreatic mass: Preliminary report. Cancer Biomark. Sect. A Dis. Markers 2016, 16, 537–543.
- Cui, Y.; Shu, X.O.; Li, H.L.; Yang, G.; Wen, W.; Gao, Y.T.; Cai, Q.; Rothman, N.; Yin, H.Y.; Lan, Q.; et al. Prospective study of urinary prostaglandin E2 metabolite and pancreatic cancer risk. Int. J. Cancer 2017, 141, 2423–2429.
- Yip-Schneider, M.T.; Soufi, M.; Carr, R.A.; Flick, K.F.; Wu, H.; Colgate, C.L.; Schmidt, C.M. Performance of candidate urinary biomarkers for pancreatic cancer—Correlation with pancreatic cyst malignant progression? Am. J. Surg. 2020, 219, 492–495.
- Debernardi, S.; O’Brien, H.; Algahmdi, A.S.; Malats, N.; Stewart, G.D.; Pljesa-Ercegovac, M.; Costello, E.; Greenhalf, W.; Saad, A.; Roberts, R.; et al. A combination of urinary biomarker panel and PancRISK score for earlier detection of pancreatic cancer: A case-control study. PLoS Med. 2020, 17, e1003489.
- Terasawa, H.; Kinugasa, H.; Ako, S.; Hirai, M.; Matsushita, H.; Uchida, D.; Tomoda, T.; Matsumoto, K.; Horiguchi, S.; Kato, H.; et al. Utility of liquid biopsy using urine in patients with pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2019, 20, 1348–1353.
- Debernardi, S.; Massat, N.J.; Radon, T.P.; Sangaralingam, A.; Banissi, A.; Ennis, D.P.; Dowe, T.; Chelala, C.; Pereira, S.P.; Kocher, H.M.; et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am. J. Cancer Res. 2015, 5, 3455–3466.
- Yoshizawa, N.; Sugimoto, K.; Tameda, M.; Inagaki, Y.; Ikejiri, M.; Inoue, H.; Usui, M.; Ito, M.; Takei, Y. miR-3940-5p/miR-8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Oncol. Lett. 2020, 19, 2677–2684.
