Nails are highly keratinized skin appendages that exhibit continuous growth under physiological conditions and full regeneration upon removal. These mini-organs are maintained by two autonomous populations of skin stem cells. The fast-cycling, highly proliferative stem cells of the nail matrix (nail stem cells (NSCs)) predominantly replenish the nail plate. Furthermore, the slow-cycling population of the nail proximal fold (nail proximal fold stem cells (NPFSCs)) displays bifunctional properties by contributing to the peri-nail epidermis under the normal homeostasis and the nail structure upon injury. Here, we discuss nail mini-organ stem cells’ location and their role in skin and nail homeostasis and regeneration, emphasizing their importance to orchestrate the whole digit tip regeneration. Such endogenous regeneration capabilities are observed in rodents and primates. However, they are limited to the region adjacent to the nail’s proximal area, indicating the crucial role of nail mini-organ stem cells in digit restoration.
- nail organ
- nail stem cells (NSCs)
- nail proximal fold stem cells (NPFSCs)
- nail matrix
- nail regeneration
- skin regeneration
- digit tip regeneration
- BMP signaling
- WNT signaling
1. Dissection of Nail Mini-Organ Anatomy and Function
Nails are the skin appendages located at the distal phalanx of each finger and toe of the human body, providing physical protection of the dorsal tips and assistance in fine manipulations and subtle finger functions, including scratching. The structure begins at the eponychium, separating a thickened layer of skin epidermis from the nail organ, which firmly adheres to the nail plate and protects the area between the nail and epidermis from exposure to germs and contamination. At this point, the skin epidermis folds inwards ventrally, forming the nail proximal fold (NPF), which is histologically different from normal skin epidermis due to a ceasing epidermal differentiation, manifested by a loss of the granular layer with loricrin expression (Figure 1) [1]. As the wedge-shaped NPF bends once more, it attaches to the root nail plate’s dorsal surface forming the nail matrix towards the digit’s tip. The thick proliferating nail epithelium zones are profusely connected with nerves and are supplied with lymph and blood vessels. These actively proliferating nail keratinocytes are called the onychocytes, which establish the eosinophilic keratogenous zone (KZ). Next, the KZ differentiates into corneocytes, forming a protective, strong nail plate (NP) (Figure 1). During the maturation process, onychocytes broaden and flatten out, depositing to the NP. Once their nuclei disintegrate, onychocytes become corneocytes with AE13-positive hard keratins’ expression. The loss of nuclei contributes to the nail plate’s translucency, allowing the nail matrix at the NP’s base to be perceived in the form of a whitish crescent-shaped area called the lunula (Figure 1). The NP’s edges are surrounded by nail grooves and lateral nail folds. The hyponychium seals the plate to the finger at the distal end border and the beginning of the volar epidermis, preventing pathogens and pollution from entering underneath the plate (Figure 1). The folds under the surface of the NP, spanning between the end of the distal nail matrix (edge of the lunula) to the hyponychium, firmly attach to the complementary longitudinal epidermal ridges of the underlying nail bed (NB) and fasten the nail plate to the surface (Figure 1) [2]. The thin epithelium of NB consists of one or two layers of suprabasal postmitotic keratinocytes.
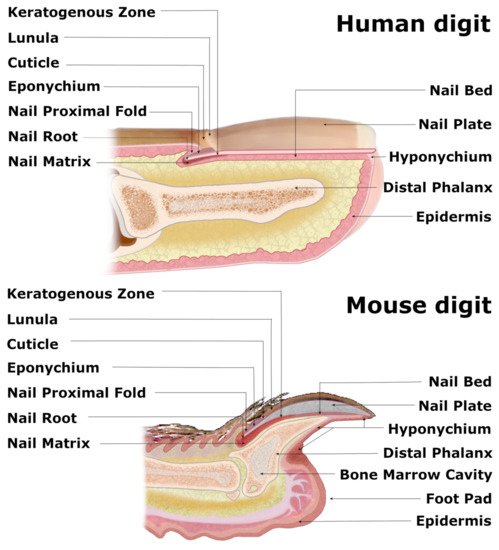
Figure 1. Comparative anatomy of human and mouse nail mini-organs. The visibly distinguishable nail organs of human and mouse digits share major characteristics across their inner structures. In both species, the wedge-shaped nail proximal fold (NPF), which smoothly transitions into the nail matrix (perceived as whitish crescent-shaped lunula), encircles the nail root of the nail plate (NP). Both flat human NP and claw-shaped murine NP lie atop of the keratogenous zone (KZ) and the nail bed (NB) and are protected with eponychium at the nail root and the hyponychium underneath the distal end of the NP. The triangular-shaped bone of the murine digit and splayed terminal phalanx of the human appendage enclose the bone marrow cavity.
The visual distinctions between the digits of different mammals are apparent, as their function and evolution dictate the shapes of the nails and claws. However, recent studies have demonstrated that these units’ inner structures share major characteristics, especially human and mouse nail organs, with the latter already successfully employed as a model for the homologous human appendage [3]. Although the longitudinally curved murine NP, terminated with a sharp tip, is visibly distinguishable from a relatively flat and round-ended human nail, both organs consist of similar structures, including the NPF with eponychium, nail matrix along with KZ, NB, NP and hyponychium [4]. The base of the ventral surface of the NPF gives rise to the thick nail matrix (Figure 2), dorsally coated with KZ (Figure 1). Both layers suddenly become thin as they merge with the NB. A thin layer of loose connective tissue, rich in fibroblasts, vasculature and nerves, separates the nail epidermis from the triangular-shaped bone. Mouse NP encircles the distal phalanx from dorsal and lateral sides while leaving the exceptionally extended hyponychium uncovered (Figure 1).
Furthermore, to both structures sharing major characteristics, most keratins’ expression patterns are similar in both human and mouse digits [3]. These resemblances make the mouse nail mini-organ a perfect model to study nail biology and investigate digit regeneration mechanisms. Moreover, a wide variety of genetically modified mice enables researchers to analyze the genetic-based nail disorders and discover ways to overcome amputated mammalian digits and limbs’ regenerative failures. Ultimately, the results obtained from studies performed on mouse models might soon translate into a potential clinical setting in humans.
2. Searching for the Nail Mini-Organ Stem Cells
For their self-renewal and differentiation ability into the various specialized cell types, the adult stem cells (SCs) are responsible for upholding the homeostasis of tissues and organs by participating in their growth, renewal and regeneration. Each organ holds a specialized microenvironment, known as the niche, which helps to maintain the quiescence and regulate the proliferation and differentiation of the inherent stem cells [5,6][5][6]. The discovery of slow-cycling or quiescent label-retaining cells (LRC) allowed for identifying several skin stem cells across the epithelium in hair follicle bulge, cornea limbus, or sweat glands [7,8,9,10,11,12,13][7][8][9][10][11][12][13]. The BrdU pulse-chase procedure has been used in mice to identify the LRCs existence in their nail organ. However, that initial discovery has incorrectly localized those slow-cycling LRCs to the nail matrix’s basal layer adjacent to the nail bed [14]. This observation stood in contradiction to discoveries based on the lineage tracing of keratin 14 positive (K14+) basal epidermal cells, labeled with LacZ, which defined the proximal nail matrix as a collection of highly proliferative keratinocytes without LRCs [15]. It was also consistent with previous studies, which used radioactive tritiated glycine to predominantly label nail matrix [16,17][16][17]. Moreover, Sellheyer and Nelson’s complementary studies [18] focused on characterizing a slowly proliferating nail cells population within the NPF. During the embryogenesis, these cells expressed the same markers as the well-known hair follicle stem cells: keratin 15 (K15), keratin 19 (K19) and pleckstrin homology like domain, family A, member 1 protein (PHLDA1) [19] (Figure 3). Thus, in the beginning, the similarity between both cell types suggested further the existence of LRCs in the nail organ, however, localized rather in the ventral proximal fold instead of the previously proposed nail matrix. Finally, the recent studies, which used the doxycycline-regulated H2B-GFP LRC system in the K5TetOffTreH2BGFP transgenic mouse, clarified these discrepancies further and managed to address the previous inconsistencies. The LRCs were confirmed to be organized in a ring-like formation around the nail root in the nail proximal fold’s basal layer [20].
A few years later, Lehoczky and Tabin [21] identified the Lgr6 receptor (leucine-rich repeat-containing G protein-coupled receptor 6), a recognized marker of several epithelial SCs populations, in the murine nail stem cells (NSCs) of the nail matrix (Figure 3) and in a small cell subset across the distal digit bone and the eccrine sweat glands within the toe pad. In contrast, the presence of Lgr5 receptors was determined only in a unique dermal population of cells adjacent to the slow-cycling LRCs in the NPF and at the distal groove of the digit (Figure 3, marked by light blue) [20,21][20][21]. However, the role of these Lgr5+ cells in nail growth and homeostasis has yet to be defined.
3. Two Pools of Nail Mini-Organ Stem Cells
Hence, two different pools of stem cell population were identified within the nail mini-organ: the highly proliferative Ki67+ cells at the proximal matrix region and the slow-cycling LRCs (H2BGFP+) in the nail proximal fold (Figure 3) [20,22][20][22]. Between both factions, a gradient of less proliferative cells, labeled by both the Ki67+ and weak H2BGFP+ expression, marked the intermediate zone (IZ). The in vivo lineage tracing experiments in transgenic K15CrePRRosa26LacZ mouse has shown that the slow-cycling K15+ nail proximal fold Stem Cells (NPFSCs) contribute to both the peri-nail epidermis and the nail plate structure, thus possessing bifunctional stem cells characteristics. Under physiological conditions, these cells are more involved in supporting the peri-nail epidermis tissue than the NP. However, following the injury, the homeostatic balance tilts toward the nail regeneration, and the NPFSCs adapt to the new circumstances by delivering the progeny to the nail matrix and differentiating into AE13-positive hard keratins of the nail plate [20].
On the other hand, the pool of NSCs discovered in the nail matrix, characterized as highly proliferative Ki67+ progenitor cells, was the main contributor to the external NP (Figure 2). These cells were located and described by Takeo et al. [15], who used a lineage tracing system in the K14CreERR26RLSL-LacZ transgenic mouse in order to mark a small subset of keratinocytes in the basal layer of skin and nail epidermis. Through a controlled expression of LacZ, the K14+ cells that took part in the nail growth were marked in the nail matrix and the nail bed. During 5 months of the experiment, the LacZ+ progenies were perceived as streaks in the NP, spreading linearly and distally from the proximal matrix. Apart from the expression of highly proliferative Ki67 marker and keratin 14 (K14), further analyses determined that proximal matrix cells expressed keratin 17 (K17) and possessed a high colony-forming ability observed in vitro. This feature confirmed that the proximal matrix indeed contained self-renewing NSCs that sustain nail growth. In comparison, no LacZ+ labeled cells were observed to emerge from the distal matrix.
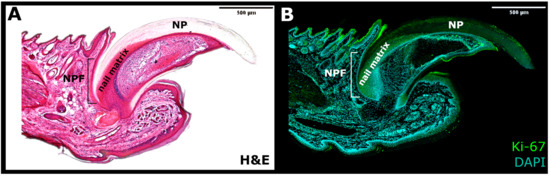
Figure 2. The side section of the nail mini-organ. (A) Haematoxylin and eosin (H&E) staining. (B) Immunofluorescence for Ki-67 marker of proliferation (Green) and DAPI (Blue). NPF—nail proximal fold; NP—nail plate.
The gene expression evaluation, performed between the distal and proximal matrix, characterized a major of the proximal NSCs with a downregulated Wnt signaling pathway, suggesting a direct involvement of the Wnt signaling in the nail differentiation [15]. The subsequently identified presence of the mediator of the Wnt signaling, the Lgr6 receptor, strongly supported this theory. Indeed, the genetic lineage-tracing experiments determined that the Lgr6+ cells contribute to the nail homeostatic growth, indicating Lgr6-marked cells as NSCs [21].
In summary, the nail mini-organ holds two distinct populations of stem cells, located in the nail matrix and the nail proximal fold. During the physiological nail growth, the matrix pool continuously delivers cells to the nail plate, while cells localized in the NPF predominantly support the peri-nail epidermis. The bifunctional characteristic of the NPFSCs becomes apparent upon a nail injury when the progenies from the NPF hasten to enrich the nail matrix and contribute to the regenerating nail plate.
4. Nail Growth, Differentiation and Homeostasis
The development of the nail organ in the human embryo begins around the 10th week of gestation [1]. At the 14th week, NP emerges from NPF and by the 17th week, the whole NB’s surface is covered by the NP. The fingernail of a healthy adult takes around 6 months to grow entirely, while this time is doubled for toenails [23]. It is generally acknowledged, that the nail matrix is predominantly accountable for the replenishing of the NP, while the bifunctional NPF cells can also contribute to the process; however, there is still an ongoing discussion whether other elements of the nail organ take part in the NP growth, especially the nail bed. A study performed by Zaias and Alvarez [16] confirmed that the nail matrix cells move superficially into the nail plate and distally into NB. The continuous replacement of the nail matrix is ensured by the cells’ divisions at its basal layer, which move the differentiating cells up, to be deposed through a KZ to the nail plate. At the same time, the pressure of accumulated cells moves the NP dorsally. However, based on the thickness and mass of the nail plate, the relatively inactive nail bed cells were also proposed to contribute a few horn cells to the distal nail plate’s ventral surface, possibly facilitating the distal movement as it grows [4,24][4][24]. Indeed, NB was observed to comprise cells expressing Msx1, a marker associated with multipotent and relatively undifferentiated progenitor cells, active within the developing and regenerating environments [25,26][25][26]. The minor input of the NPFSCs to the NP during the normal nail homeostasis was described by Leung et al. [20]. Further, the authors investigated these slow-cycling cells’ responsiveness upon a mechanical NP plucking on the K15CrePRRosa26Tom mouse at the postnatal day 7 (P7). The keratinized structure entirely grew back within 2 weeks. The linear streaks of Tom+ cells (marked by Tomato) confirmed that the K15+ NPFSCs actively delivered progenies to the nail matrix and differentiated into the NP. NPFSCs also participated in the elongation of the NPF, providing extended protection of the exposed tissue. The following engraftment procedure of the H2BGFP+ nail LRC strip directly underneath the NPF of the immunocompromised NOD SCID donors confirmed the regenerative ability of the NPF LRCs [20]. Within several days, the engrafted cells were observed to successfully integrate with NPFSCs and contribute to the growing nail while retaining their slow-cycling physiology after an additional chase period. These findings established that normally quiescent NPFSCs are bifunctional and are able to adapt to the changing environmental circumstances by contributing to the nail restoration upon injury.
5. Digit Regeneration
Tissue regeneration is a complex process that aims to re-establish the polarity, structure and functionality of the disturbed or damaged fragment of the organ [37][27]. The digit tips of rodents and primates are recognized for their endogenous regeneration capability, including the digit’s nail plate, epidermis, nerves and bone. Nevertheless, this capacity was observed to be limited to the region adjacent to the nail organ area [38,39][28][29]. The distal amputation, which ensures consequent complete regeneration, removes around 23% of the distal phalanx length and 15% of the bone volume but leaves the bone marrow (BM), proximal nail matrix and footpad intact [40][30]. The extended amputation executed past this proximal boundary does not trigger an analogous regenerative response (Figure 3). This observation indicates that the nail epithelium and particularly the NSCs of the proximal nail matrix have a crucial role in orchestrating the complete digit tip regeneration.
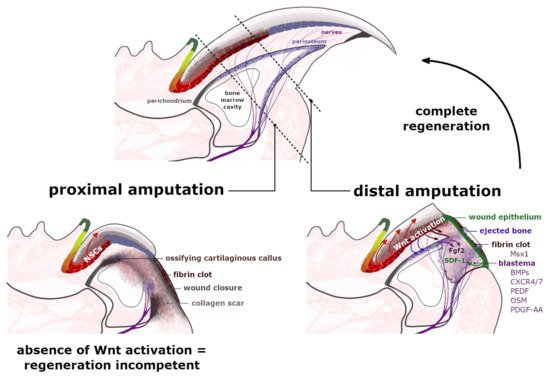
Regeneration of the amputated digit tips. Proximal amputation eliminates over 50% of the distal phalanx, removing the distal region of matrix epithelium, the periosteum, share of the bone marrow and the footpad while leaving the nails stem cells (NSCs) of the proximal matrix intact. Following the fibrin clot formation and the wound closure at the amputation level, a cartilaginous callus ossifies annularly along the bone surface, broadening the shortened stump along its dorsal-ventral axis. The formation of a dense collagen scar leaves the injured site regenerative-incompetent. The distal amputation, which eliminates up to 23% of the distal phalanx length, initiates the histolysis of the following 30% of the remaining stump. Between the ejected bone, protruded in a fibrin clot and an exposed bone marrow cavity, a transient wound epidermis (WE) forms, which through secretion of SDF-1 (green arrow) acts as a signaling center for CXCR4/7
and Pedf
cells. The WE outlines the blastema—an accumulation of highly proliferative cells, which express the bone morphogenetic proteins (BMPs) and antiangiogenic factor PEDF and ultimately contribute to the regenerating structures. The Msx1
cells, present in the fibrin clot, do not build the blastema. The expression of Wntless protein by a distal nail matrix activates the Wnt signaling pathway, which promotes blastema innervation (burgundy arrows), which in turn provides the Fgf2 expression (purple arrows). Additionally, injured peripheral nerves provide a migration route for the Schwann cell precursors that secrete additional growth factors, including Oncostatin M (OSM) and platelet-derived growth factor AA (PDGF-AA). The presence of growth factors stimulates blastema cells to proliferate and differentiate. Furthermore, the active Wnt signaling interacts with periosteum (blue arrows) containing Wnt-responsive mesenchymal cells, which are crucial for the distal appositional bone growth and nail bed extension.
The regeneration of an amputated digit tip is a multistep process, which commences with an inflammation of the damaged tissue as it becomes infiltrated by macrophages and neutrophils [41][31]. At this point, as the blood vessels are opened, the soft tissue of the injured site swells and a fibrin clot forms over the cut area [25]. During the following phase, called histolysis, the extracellular matrix, including the bone’s surface, is enzymatically degraded and erodes into two segments, ultimately exposing the bone marrow [39][29]. Accountable for this erosion response are the osteoclasts, which were observed to increase their volume directly after the amputation and during the histolysis process. The timing of the bone degradation in mice, studied by Fernando et al. [42][32], varied between 9–12 days following the injury [43][33] and strongly related to the completion of the wound closure, which was visibly inhibited by the remaining stump bone [44][34]. At the beginning of the histolysis process, the digit’s upper epidermis attaches to the lateral rim of the bone at the amputation level. Simultaneously, the ventral site’s tissue retracts proximally to connect to the external bone surface at the border of the emerging bone marrow cavity. The progressive degradation of roughly 50% of the distal phalanx bone volume provides a migration route for the epidermal cells. Newly formed transient wound epidermis (WE) separates the eroded distal bone stump, protruded into the clot (Figure 3). The WE is required for the digit blastema to establish and develop. For the secreted chemoattractant, stromal cell-derived factor 1 (SDF-1), WE acts as a signaling center for blastema cells. These cells are recognized to predominantly express the chemokine receptors type 4 and 7 (CXCR4/7) and pigment-epithelium derived factor (PEDF) receptors for the SDF-1 (Figure 3) [40,45][30][35]. Additionally, apart from facilitating cell migration, SDF-1 also mediates cell adhesion and survival [46][36].
The wound closure, which is now proximal to the original level of the amputation, outlines the blastema—an avascular [47][37], but innervated structure, which expanses from the exposed bone marrow cavity; the source of BM-derived stem cells and bone progenitor cells [40,42][30][32]. The origin of many blastema mesenchymal cells is still in debate, but their sources are being gradually uncovered [48][38]. The latest lineage tracing analysis determined that 26% of blastema cells originate from bone-associated cells, including the periosteum [40[30][39],49], as they express the Dmp1 (Dentin matrix acidic phosphoprotein1) marker for osteoblasts and osteocytes [50][40]. The connective tissue (CT) unquestionably plays a critical role in appendage regeneration by providing many cells to the blastema and by concealing the positional identity of the limbs [51][41]. The synthesis of the extracellular matrix (ECM) of the CTs is supplied by stromal mesenchymal progenitors (MPs), essential in modeling and/or remodeling the tissues’ structural integrity. Although the MPs display functional specialization, based on their tissue of origin and localization, it was long suspected that different types of MPs are able to convert into or replace each other. The activation of the MPs marker PDGF-α (platelet-derived growth factor receptor α), triggers several cellular responses, including cell migration, proliferation and differentiation. The CT-derived PDGF signaling was indeed identified in blastema during axolotl limb regeneration [52][42]. As for mammals, the relevance of Pdgfra+ MPs was also determined in the tissue restoration processes [53,54][43][44]. Most recent studies performed by Storer et al. [50][40] exposed that blastema is mainly formed by Pdgfra+ cells and that their progeny locate throughout the regenerating bone and dermis (Figure 3). Ultimately, ablation of the Pdgfra+ cells in the mouse model compromised the regeneration of the digit.
Several hypotheses were formed in order to explain the following regenerative mechanisms and determine the possible sources of the blastema cells. It was previously believed that these highly proliferative cells are all lineage-restricted, thus, are only the progenitors of their own tissue of origin [55,56][45][46]. Many challenged this concept, including Carr et al. [54][44], who studied nerve-derived mesenchymal cells in the blastema and identified their descendants in the dermis and bone of the regenerated digit. Other theories assumed the recruitment of the circulating precursors, which would transdifferentiate within the damaged tissue [52,57,58][42][47][48] and/or the involvement of the residual mature cells, which thenceforth dedifferentiated in the blastema [25,59,60,61,62,63][25][49][50][51][52][53]. The accumulation of such pluripotent class cells would further respond to different signaling pathways—other than those used in the development of digits and limbs. Indeed, it is currently under deliberation whether the blastema’s local environment affects the recruited cells’ phenotypes and, therefore, their contribution to the regenerating tissues. The single-cell RNA sequencing techniques (scRNA-seq), utilized by Storer et al. [50][40], provided new insights into the behavior, plasticity and heterogeneity of the CT progenitor cells within the blastema. The Pdgfra+ MPs were observed to acquire a unique plastic transcriptome that progresses and adapts over time as the blastema matures (Figure 3). The authors observed that the bone-lining Dmp1+ cells, in response to the digit tip injury, were capable of switching their mesenchymal lineage fate by depositing to both the bone and the dermis. Concurrently, the phenotypes of the dermal fibroblasts that were transplanted into the regenerating digits were considerably affected by the blastema environment and began to express the blastema-state genes. Ultimately, the injected fibroblasts contributed to the renewal of the bone. The same transplantation, performed in the proximally amputated, therefore non-regenerative digits, had no similar effect on the phenotype of the introduced cells, confirming the role of blastema in altering the inherent cells’ transcriptional state [50][40]. Cell population in the early blastema (11–17 days post-amputation) of the regenerating digit tip was recently analyzed via scRNA-seq by Johnson et al. [64][54], who identified the very same broad cell types in the intact unamputated digit tip. The most abundant and heterogeneous population within the blastema were fibroblasts, which were also observed to be enriched in several regeneration-specific markers, including a novel Mest gene [64][54]. The Mest gene expression was previously identified in mesenchymal tissues of the developing embryo [65][55].
In the early healing stages, the bone-forming regions are located alongside the periosteal surface of the remaining stump and are rich in actively proliferating cells that express markers for osteoblast commitment: Runx2, Sp7 and Osterix (OSX) [15,25,39,40][15][25][29][30]. The essential role of the periosteum in a successful digit tip regeneration was recognized by Dawson et al. [40][30], who showed that the tissue’s mechanical removal leads to a substantial reduction of the regenerated bone’s length and volume. Prior to the WE formation, the area connected with the periosteum forms a circumferential ring around the degrading bone. Only after the region merges with the endosteal/marrow compartment, the blastema proper is formed. At this point, the bone re-differentiation occurs, which is perceived as an ossification occurring distally to the stump [40][30]. Cells that participate in the process organize themselves through the extracellular matrix production, rich in Collagen III, which is subsequently degraded and replaced as the newly regenerated structures develop [39,66][29][56]. The process of ECM degradation and remodeling is promoted by matrix metalloproteinases (MMPs), activated by mechanical and chemical stimuli, triggered by the injury [67][57]. Initially, the restored bone, with an excessive volume at the dorsal-ventral axis, is rather trabecular in its nature, but ultimately it regains the general anatomy, length and tapered morphology of the native distal phalanx [42,44][32][34].
The healing process of the proximally amputated digit commences similarly to the regenerative response—with the inflammation and partial bone histolysis. However, the bone degradation is not as consistent and depends upon amputation level. Subsequently, cells that form the WE migrate across the top of the stump bone, separating the fibrin clot and completing the re-epithelialization within 13 days post-amputation [43][33]. At this point, fibroblastic cells, localized at the bone’s distal end, cover the stump with collagen fibrils and the cartilaginous callus forms annularly along the periosteal surface. Simultaneously, a dense collagen scar is formed in front of the bone stump. The callus’ ossification ultimately leaves the bone wider but shorter in length compared to the original amputation level. The Pdgfra+ MPs previously recognized to play a crucial role in blastema formation were also found in the non-regenerative environment and were observed to partially express blastema-associated genes [50][40]. However, these cells only contributed to the renewal of the bone stump. These results suggest that the inherent Pdgfra+ cells prime to partake in the regeneration but lack the decisive cues and signaling pathways that would reveal their plasticity and broad lineage potential.
References
- Saito, M.; Ohyama, M.; Amagai, M. Exploring the biology of the nail: An intriguing but less-investigated skin appendage. J. Dermatol. Sci. 2015, 79, 187–193.
- Brahs, B.; Bolla, S.R. Histology, Nail; StatPearls Publishing LLC: Treasure Island, FL, USA, 2019.
- Fleckman, P.; Jaeger, K.; Silva, K.A.; Sundberg, J.P. Comparative anatomy of mouse and human nail units. Anat. Rec. 2013, 296, 521–532.
- Kobielak, K. Fitzpatrick’s dermatology in general medicine. In Fitzpatrick’s Dermatology in General Medicine, 9th ed.; McGraw-Hill Education: New York, NY, USA, 2019; Chapter 8; pp. 106–115.
- Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. A hypothesis. Blood Cells 2021, 4, 7–25. Available online: (accessed on 27 January 2021).
- Daszczuk, P.; Mazurek, P.; Pieczonka, T.D.; Olczak, A.; Boryń, L.M.; Kobielak, K. An intrinsic oscillation of gene networks inside hair follicle stem cells: An additional layer that can modulate hair stem cell activities. Front. Cell Dev. Biol. 2020, 8, 595178.
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 1989, 57, 201–209.
- Cotsarelis, G.; Sun, T.-T.; Lavker, R.M. Label-Retaining Cells Reside in the Bulge Area of Pilosebaceous Unit: Implications for Follicular Stem Cells, Hair Cycle, and Skin Carcinogenesis. Cell 1990, 61, 1329–1337.
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the epithelial stem cell niche in skin. Science 2004, 303, 359–363.
- Lu, C.P.; Polak, L.; Rocha, A.S.; Pasolli, H.A.; Chen, S.-C.; Sharma, N.; Blanpain, C.; Fuchs, E. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 2012, 150, 136–150.
- Leung, Y.; Kandyba, E.; Chen, Y.B.; Ruffins, S.; Kobielak, K. Label retaining cells (LRCs) with myoepithelial characteristic from the proximal acinar region define stem cells in the sweat gland. PLoS ONE 2013, 8, e74174.
- Kobielak, K.; Kandyba, E.; Leung, Y. Skin and skin appendage regeneration. In Translational Regenerative Medicine; Academic Press: Cambridge, MA, USA, 2015; pp. 269–292.
- Morris, R.J.; Liu, Y.; Marles, L.; Yang, Z.; Trempus, C.; Li, S.; Lin, J.S.; Sawicki, J.A.; Cotsarelis, G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 2004, 22, 411–417.
- Nakamura, M.; Ishikawa, O. The localization of label-retaining cells in mouse nails. J. Investig. Dermatol. 2008, 128, 728–730.
- Takeo, M.; Chou, W.C.; Sun, Q.; Lee, W.; Rabbani, P.; Loomis, C.; Taketo, M.M.; Ito, M. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature 2013, 499, 228–232.
- Zaias, N.; Alvarez, J. The formation of the primate nail plate. An autoradiographic study in squirrel monkey. J. Investig. Dermatol. 1968, 51, 120–127.
- Norton, L.A. Incorporation of thymidine-methyl-H3 and glycine-2-H3 in the nail matrix and bed of humans. J. Investig. Dermatol. 1971, 56, 61–68.
- Sellheyer, K.; Nelson, P. The ventral proximal nail fold: Stem cell niche of the nail and equivalent to the follicular bulge—A study on developing human skin. J. Cutan. Pathol. 2012, 39, 835–843.
- Kalmukova, O. Stem cells in nail unit of mammalians. Cell Organ Transpl. 2016, 4, 138–143.
- Leung, Y.; Kandyba, E.; Chen, Y.B.; Ruffins, S.; Chuong, C.M.; Kobielak, K. Bifunctional ectodermal stem cells around the nail display dual fate homeostasis and adaptive wounding response toward nail regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 15114–15119.
- Lehoczky, J.A.; Tabin, C.J. Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc. Natl. Acad. Sci. USA 2015, 112, 13249–13254.
- Shi, J.; Lv, Z.; Nie, M.; Lu, W.; Liu, C.; Tian, Y.; Li, L.; Zhang, G.; Ren, R.; Zhang, Z.; et al. Human nail stem cells are retained but hypofunctional during aging. J. Mol. Histol. 2018, 49, 303–316.
- Baswan, S.; Kasting, G.B.; Li, S.K.; Wickett, R.; Adams, B.; Eurich, S.; Schamper, R. Understanding the formidable nail barrier: A review of the nail microstructure, composition and diseases. Mycoses 2017, 60, 284–295.
- Johnson, M.; Shuster, S. Continuous formation of nail along the bed. Br. J. Dermatol. 1993, 128, 277–280.
- Lehoczky, J.A.; Robert, B.; Tabin, C.J. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20609–20614.
- Allan, C.H.; Fleckman, P.; Fernandes, R.J.; Hager, B.; James, J.; Wisecarver, Z.; Satterstrom, F.K.; Gutierrez, A.; Norman, A.; Pirrone, A.; et al. Tissue response and Msx1 expression after human fetal digit tip amputation in vitro. Wound Repair Regen. 2006, 14, 398–404.
- Dolan, C.P.; Dawson, L.A.; Muneoka, K. Digit tip regeneration: Merging regeneration biology with regenerative medicine. Stem Cells Transl. Med. 2018, 7, 262–270.
- Sensiate, L.A.; Marques-Souza, H. Bone growth as the main determinant of mouse digit tip regeneration after amputation. Sci. Rep. 2019, 9, 9720.
- Dawson, L.A.; Schanes, P.P.; Marrero, L.; Jordan, K.; Brunauer, R.; Zimmel, K.N.; Qureshi, O.; Imholt, F.M.; Falck, A.R.; Yan, M.; et al. Proximal digit tip amputation initiates simultaneous blastema and transient fibrosis formation and results in partial regeneration. Wound Repair Regen. 2021, 29, 196–205.
- Dawson, L.A.; Schanes, P.P.; Kim, P.; Imholt, F.M.; Qureshi, O.; Dolan, C.P.; Yu, L.; Yan, M.; Zimmel, K.N.; Falck, A.R.; et al. Blastema formation and periosteal ossification in the regenerating adult mouse digit. Wound Repair Regen. 2018, 26, 263–273.
- Simkin, J.; Sammarco, M.C.; Marrero, L.; Dawson, L.A.; Yan, M.; Tucker, C.; Cammack, A.; Muneoka, K. Macrophages are required to coordinate mouse digit tip regeneration. Development 2017, 144, 3907–3916.
- Fernando, W.A.; Leininger, E.; Simkin, J.; Li, N.; Malcom, C.A.; Sathyamoorthi, S.; Han, M.; Muneoka, K. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev. Biol. 2011, 350, 301–310.
- Simkin, J.; Han, M.; Yu, L.; Yan, M.; Muneoka, K. The mouse digit tip: From wound healing to regeneration. Methods Mol. Biol. 2013, 1037, 419–435.
- Simkin, J.; Sammarco, M.C.; Dawson, L.A.; Tucker, C.; Taylor, L.J.; van Meter, K.; Muneoka, K. Epidermal closure regulates histolysis during mammalian (Mus) digit regeneration. Regeneration 2015.
- Lee, J.; Marrero, L.; Yu, L.; Dawson, L.A.; Muneoka, K.; Han, M. SDF-1α/CXCR4 signaling mediates digit tip regeneration promoted by BMP-2. Dev. Biol. 2013, 382, 98–109.
- Ratajczak, M.Z.; Zuba-Surma, E.; Kucia, M.; Reca, R.; Wojakowski, W.; Ratajczak, J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia 2006, 20, 1915–1924.
- Yu, L.; Yan, M.; Simkin, J.; Ketcham, P.D.; Leininger, E.; Han, M.; Muneoka, K. Angiogenesis is inhibitory for mammalian digit regeneration. Regeneration 2014, 1, 33–46.
- Storer, M.A.; Miller, F.D. Cellular and molecular mechanisms that regulate mammalian digit tip regeneration. Open Biol. 2020, 10, 200194.
- Dawson, L.A.; Yu, L.; Yan, M.; Marrero, L.; Schanes, P.P.; Dolan, C.; Pela, M.; Petersen, B.; Han, M.; Muneoka, K. The periosteal requirement and temporal dynamics of BMP2-induced middle phalanx regeneration in the adult mouse. Regeneration 2017, 4, 140–150.
- Storer, M.A.; Mahmud, N.; Karamboulas, K.; Borrett, M.J.; Yuzwa, S.A.; Gont, A.; Androschuk, A.; Sefton, M.V.; Kaplan, D.R.; Miller, F.D. Acquisition of a unique mesenchymal precursor-like blastema state underlies successful adult mammalian digit tip regeneration. Dev. Cell 2020, 52, 509–524.e9.
- Tanaka, E.M. The molecular and cellular choreography of appendage regeneration. Cell 2016, 165, 1598–1608.
- Currie, J.D.; Kawaguchi, A.; Traspas, R.M.; Schuez, M.; Chara, O.; Tanaka, E.M. Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev. Cell 2016, 39, 411–423.
- Johnston, P.W.; Yuzwa, S.A.; Carr, M.J.; Mahmud, N.; Storer, M.A.; Krause, M.P.; Jones, K.; Paul, S.; Kaplan, D.R.; Miller, F.D. Dedifferentiated schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 2016, 19, 433–448.
- Carr, M.J.; Toma, J.S.; Johnston, A.P.W.; Steadman, P.E.; Yuzwa, S.A.; Mahmud, N.; Frankland, P.W.; Kaplan, D.R.; Miller, F.D. Mesenchymal precursor cells in adult nerves contribute to mammalian tissue repair and regeneration. Cell Stem Cell 2019, 24, 240–256.e9.
- Wagers, J.; Weissman, I.L. Plasticity of adult stem cells. Cell 2004, 116, 639–648.
- Rinkevich, Y.; Lindau, P.; Ueno, H.; Longaker, M.T.; Weissman, I.L. Germ-layer and lineage-restricted stem/ progenitors regenerate the mouse digit tip. Nature 2011, 476, 409–413.
- Zaidi, N.; Nixon, A.J. Stem cell therapy in bone repair and regeneration. Ann. N. Y. Acad. Sci. 2007, 1117, 62–72.
- Djouad, F.; Bouffi, C.; Ghannam, S.; Noël, D.; Jorgensen, C. Mesenchymal stem cells: Innovative therapeutic tools for rheumatic diseases. Nat. Rev. Rheumatol. 2009, 5, 392–399.
- Tamura, K.; Ohgo, S.; Yokoyama, H. Limb blastema cell: A stem cell for morphological regeneration. Dev. Growth Differ. 2010, 52, 89–99.
- Tsonis, P.A. Stem cells and blastema cells. Curr. Stem Cell Res. Ther. 2008, 3, 53–54.
- Christen, B.; Robles, V.; Raya, M.; Paramonov, I.; Belmonte, J.C.I. Regeneration and reprogramming compared. BMC Biol. 2010, 8, 5.
- Gerber, T.; Murawala, P.; Knapp, D.; Masselink, W.; Schuez, M.; Hermann, S.; Gac-Santel, M.; Nowoshilow, S.; Kageyama, J.; Khattak, S. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 2018, 362, 6413.
- Wagner, I.; Wang, H.; Weissert, P.M.; Straube, W.L.; Shevchenko, A.; Gentzel, M.; Brito, G.; Tazaki, A.; Oliveira, C.; Sugiura, T.; et al. Serum proteases potentiate BMP-induced cell cycle re-entry of dedifferentiating muscle cells during newt limb regeneration. Dev. Cell 2017, 40, 608–617.e6.
- Johnson, G.L.; Masias, E.J.; Lehoczky, J.A. Cellular heterogeneity and lineage restriction during mouse digit tip regeneration at single-cell resolution. Dev. Cell 2020, 52, 525–540.e5.
- Kaneko-Ishino, T.; Kuroiwa, Y.; Miyoshi, N.; Kohda, T.; Suzuki, R.; Yokoyama, M.; Viville, S.; Barton, S.C.; Ishino, F.; Surani, M.A. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 1995, 11, 52–59.
- Marrero, L.; Simkin, J.; Sammarco, M.; Muneoka, K. Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration 2017, 4, 69–84.
- Mu, X.; Bellayr, I.; Pan, H.; Choi, Y.; Li, Y. Regeneration of soft tissues is promoted by MMP1 treatment after digit amputation in mice. PLoS ONE 2013, 8, e59105.
