The import and export of proteins requires interaction with the components of the nuclear envelope (NE), which is formed by a double membrane that harbor protein channels called nuclear pore complexes (NPCs). NPCs are formed by multiple copies of Nups that participate in the bi-directional nucleus–cytoplasmic transport of macromolecules, ribosomal subunits, viral proteins and RNAs (mRNAs, rRNAs, tRNAs, miRNAs) from both cellular and viral origin.
- nuclear pore complex
- nucleus
- nucleoporin
- nuclear localization sequence
1. ThNPC Structure Machinery of The Nuclear Pore Complex (NPC)and Function in the Cell: One Way to Access the Nucleus
1.1. The Machinery of The Nuclear Pore Complex (NPC)
The import and export of proteins requires interaction with the components of the nuclear envelope (NE), which is formed by a double membrane that harbor protein channels called nuclear pore complexes (NPCs). NPCs are formed by multiple copies of Nups that participate in the bi-directional nucleus–cytoplasmic transport of macromolecules, ribosomal subunits, viral proteins and RNAs (mRNAs, rRNAs, tRNAs, miRNAs) from both cellular and viral origin [1][2][3].
The NPC is made up of approximately 30 multi-copy of Nups, arranged from the cytoplasm to the nucleoplasm, which makes up the cytoplasmic filaments, cytoplasmic ring, internal pore ring, nuclear ring, and nuclear basket [4][5]. Inside the NPC, there are Nups with repeating sequences rich in phenylalanine (Phe) and glycine (Gly), called FG-Nups, such as Nup358, Nup62, Nup58, Nup54, Nup98, Nup45, Nup214, hCG1, Nup153, TPR, and Nup50, that facilitate the nuclear transport in both directions since they join to nuclear transport receivers (NTRs) (Figure 1) [6][7][8].
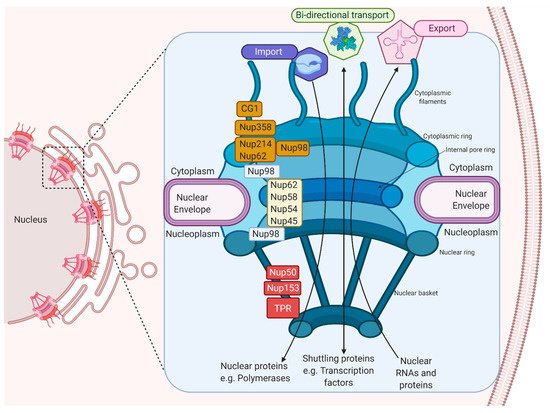
Figure 1. The machinery of the Nuclear Pore Complex. The NPC’s different components, which participate in the import and export of proteins and RNAs, and the FG-Nups, which participate in nuclear–cytoplasmic transport, are shown.
2. Bidirectional Nucleus–Cytoplasm Transport

Figure 1. The machinery of the Nuclear Pore Complex. The NPC’s different components, which participate in the import and export of proteins and RNAs, and the FG-Nups, which participate in nuclear–cytoplasmic transport, are shown.
1.2. Bidirectional Nucleus–Cytoplasm Transport
Molecules smaller than approximately 40–50 kDa can pass freely through the nuclear envelope; however, higher molecular weight molecules such as proteins and RNAs from both cellular and viral origin are actively transported through the NPC, between the nucleoplasm and the cytoplasm. The nuclear import and export of molecules are regulated by NTR (nuclear transport receptors), such as importins, exportins, carriers, and small GTPases of the Ran family that regulate the activity of importins and exportins that transport cargo molecules (Figure 2) [8][9][10].
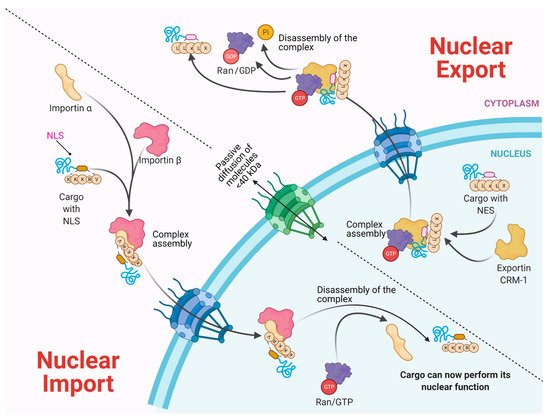
Figure 2.

Figure 2. Bidirectional Nucleus–cytoplasm Transport. In the classical pathway of import (importin α/β), the importins and cargo complex are formed through the recognition of the NLS (nuclear location sequence). In the case of Export via CRM-1, the charge contains an NES (nuclear export sequence). In both cases, a GTP gradient is required. Molecules <40 kDa pass through passive diffusion towards the nucleus by the NPC.
NTRs recognize specific sequences in cargo proteins that cross the nuclear membrane from the cytoplasm, such as nuclear location sequences (NLS) that contain repeated arginine (Arg or R) and lysine (Lys or K) amino acids. The classical NLS consists of five KKKRK amino acids. Moreover, some proteins possess bipartite NLS consisting of two groups of basic amino acids, separated by approximately ten amino acids [11][12]. On the other hand, nuclear export sequences (NES) participate in the trafficking from the nucleus to the cytoplasm. They are composed of sequences rich in leucine or hydrophobic amino acids such as valine (Val), isoleucine (Ile), phenylalanine (Phe), or methionine (Met), which are found in motifs conserved in cargo proteins, such as some transcription or translation factors and mRNA transport proteins [13][14][15]. The nuclear localization of a given protein occurs by the recognition of the NLS by the NTRs; for example, importin α through its NTR binds to NLS-cargo, then importin β binds to importin α to form a trimeric complex and its cargo molecule. If the NLS is atypical, then importin β directly binds to its cargo molecule without the participation of importin α. The directionality of the cargo is given by the Ran-GTP gradient, regulated by the Ran-GTP/GDP cycle. Once the trimeric complex has entered the nucleus, the Ran-GTP activated by RCC1 (GEF) joins importin β and thereby is released from its cargo. Importin β is transported to the cytoplasm, and the Ran-GTP is deactivated to Ran-GDP by GAP (GTPase Activating Protein) to free itself from importin β for its next import cycle.
On the other hand, the nuclear export is given by the recognition of the nuclear export sequences (NES). Nuclear export begins with Ran-GTP binding to exportin (e.g., CRM-1), which causes an increased affinity for the export cargo. Then, the complex moves to the nuclear pore and Ran-GTP hydrolyze (activated by RCC1), which forms the export complex. The complex crosses the NPC, and in the cytoplasm, GAP deactivates RanGTP (hydrolyses the GTP in GDP), causing the export protein to be released from its cargo [11][12][16].
References
- Beck, M.; Hurt, E. The Nuclear Pore Complex: Understanding Its Function through Structural Insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89.
- Hezwani, M.; Fahrenkrog, B. The Functional Versatility of the Nuclear Pore Complex Proteins. Semin. Cell Dev. Biol. 2017, 68, 2–9.
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The Human Nuclear Pore Complex as Revealed by Cryo-Electron Tomography. Structure 2012, 20, 998–1006.
- Jahed, Z.; Soheilypour, M.; Peyro, M.; Mofrad, M.R.K. The LINC and NPC Relationship—It’s Complicated! J. Cell Sci. 2016, 129, 3219–3229.
- Schwartz, T.U. The Structure Inventory of the Nuclear Pore Complex. J. Mol. Biol. 2016, 428, 1986–2000.
- Li, C.; Goryaynov, A.; Yang, W. The Selective Permeability Barrier in the Nuclear Pore Complex. Nucleus 2016, 7, 430–446.
- Rothballer, A.; Kutay, U. SnapShot: The Nuclear Envelope I. Cell 2012, 150, 868.
- Wente, S.R.; Rout, M.P. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb. Perspect. Biol. 2010, 2, a000562.
- Jovanovic-Talisman, T.; Zilman, A. Protein Transport by the Nuclear Pore Complex: Simple Biophysics of a Complex Biomachine. Biophys. J. 2017, 113, 6–14.
- Kabachinski, G.; Schwartz, T.U. The Nuclear Pore Complex—Structure and Function at a Glance. J. Cell Sci. 2015, 128, 423–429.
- Christie, M.; Chang, C.-W.; Róna, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.S.; Fontes, M.R.M.; Stewart, M.; Vértessy, B.G.; Forwood, J.K.; et al. Structural Biology and Regulation of Protein Import into the Nucleus. J. Mol. Biol. 2016, 428, 2060–2090.
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six Classes of Nuclear Localization Signals Specific to Different Binding Grooves of Importin α. J. Biol. Chem. 2009, 284, 478–485.
- Hutten, S.; Kehlenbach, R.H. CRM1-Mediated Nuclear Export: To the Pore and Beyond. Trends Cell Biol. 2007, 17, 193–201.
- La Cour, T.; Gupta, R.; Rapacki, K.; Skriver, K.; Poulsen, F.M.; Brunak, S. NESbase Version 1.0: A Database of Nuclear Export Signals. Nucleic Acids Res. 2003, 31, 393–396.
- Xu, D.; Farmer, A.; Chook, Y.M. Recognition of Nuclear Targeting Signals by Karyopherin-β Proteins. Curr. Opin. Struct. Biol. 2010, 20, 782–790.
- Musso, D.; Gubler, D. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524.
