MM is a hematological neoplasm that is still considered an incurable disease. Besides established genetic alterations, recent studies have shown that MM pathogenesis is also characterized by epigenetic aberrations, such as the gain of de novo active chromatin marks in promoter and enhancer regions and extensive DNA hypomethylation of intergenic regions, highlighting the relevance of these non-coding genomic regions. A recent study described how long non-coding RNAs (lncRNAs) correspond to 82% of the MM transcriptome and an increasing number of studies have demonstrated the importance of deregulation of lncRNAs in MM.
- lncRNAs
- multiple myeloma
- RNA-based therapy
1. Introduction
Multiple myeloma (MM) is a hematological neoplasm characterized by the uncontrolled aberrant clonal proliferation of plasma cells (PCs) in the bone marrow [1]. This disease is the second most common hematological malignancy, after non-Hodgkin lymphoma [2][3], affecting elderly patients with a median age of 65 years [4]. Despite the latest advances in treatment strategies, which have significantly increased patient survival, MM is still considered an incurable disease, with a median overall survival of 7 years.
MM is a very heterogeneous disease, which is reflected in the inter-individual differential diagnosis and survival of patients. Different studies have associated this variability with a wide range of genetic and epigenetic alterations present in MM patients [5][6], including distinct molecularly defined subtypes with different features [7]. Regarding the genetic variability, MM is divided into hyperdiploid (HRD) and non-HRD subtypes [7]. HRD MM is characterized by the trisomy of chromosomes 3, 5, 7, 9, 11, 15, 19 and 21 [6], whereas non-HRD MM is characterized by translocations of the immunoglobulin (Ig) alleles. The majority of these translocations affect chromosome 14, where the Ig H-chain is located [6][7]. However, Ig translocations can also affect the kappa and lambda light chains, the co-occurrence of which is common with HRD MM. Besides, some of these light-chain translocations are associated with a poor outcome for MM patients, as is the case for IgL-MYC translocations [8]. Some of the common heavy-chain translocations are also considered as high-risk prognostic factors, such as t(4;14) and t(14;16), which affect
MMSET
MAF genes, respectively [6][7][9]. Epigenetic aberrations of the DNA methylation and histone modifications are also thought to play an important role in MM pathogenesis. The study of global DNA methylation of MM has led to the identification of a highly heterogeneous DNA methylation pattern, which results in extensive DNA hypomethylation of intergenic regions and DNA hypermethylation associated with intronic and enhancer regions [2][5]. In addition, the study of histone modifications in MM has revealed a de novo gain of active chromatin marks preferentially located in regulatory elements, such as enhancer and promoter regions, which arise from heterochromatic regions in normal B cells [10][11][12]. These results suggest the possibility that these epigenetically regulated non-coding genomic regions could lead to the transcription of non-coding RNA genes (ncRNAs) and, in particular, to the expression of long non-coding RNAs (lncRNAs), which may play a relevant role in the pathobiology and clinical outcome of MM [13]. Nowadays, studies about the role of certain lncRNAs in MM are emerging. However, more comprehensive analyses are required to better understand their function in this disease. In this review, we summarize the current knowledge regarding the role of lncRNAs in the development and outcome of MM and discuss the possibility of lncRNAs as targets for the development of novel RNA-based therapeutic strategies for MM patients.
2. Features of lncRNAs
Traditionally, cellular functions of DNA and proteins have overshadowed the roles of RNAs. In recent years, the development of high-throughput techniques, such as RNA sequencing (RNA-seq), has brought great advances in the understanding of the cell transcriptome. So far, it is known that, although only 1–2% of the human genome is translated into proteins, around 70–90% of it is transcribed into RNA, resulting in a huge amount of ncRNAs [14]. Among these ncRNA genes, lncRNAs are defined as those non-coding transcripts longer than 200 nt that do not encode proteins, with open reading frames (ORFs) smaller than 100 amino acids, and with a lack of or low coding potential. However, the latest RNA-seq studies have shown that some lncRNAs contain cryptic ORFs, which could encode for small ORFs or non-conserved peptides [15][16][17].
MALAT1
NEAT1 (nuclear paraspeckle assembly transcript 1) [18][19]. Although the size of lncRNAs varies between 200 nt and more than 1 MB (known as macro lncRNAs), 42% of lncRNAs only present two exons [19][20]. In contrast to mRNAs, which are located at the cytosol, lncRNAs can be located either in the nucleus or in the cytoplasm, where they can exert various functions. Thus, regarding the location where lncRNAs act and their transcription site, they are capable of acting as
cis
Cis
trans lncRNAs act over distal genes [22][23][24]. Interestingly, lncRNAs are cell- and tissue-specific, and they may affect different biological processes, such as chromosome conformation, imprinting of genomic loci, or gene and protein regulation [15][25]. lncRNAs have the ability to regulate at DNA, RNA and protein levels, and their functions can be divided into four different groups depending on their molecular mechanisms [18][26]: (1) signal lncRNAs are regulatory molecules that can trigger the transcription of other genes by their presence. They can infer chromatin states, affect gene imprinting or mark certain spaces, times or stages for gene regulation, such as
Air
PANDA (p21-associated ncRNA DNA damage activated) [26][27]. (2) Decoy lncRNAs are transcripts that bind to targets and prevent them from binding to their own targets, thus leading to the alteration of post-transcriptional control. This type of lncRNA can act as an miRNA sponge, binding to miRNAs thanks to their complementary sequence (
PTENP1 (phosphatase and tensin homolog pseudogene 1), for example, leads to tumor suppressor activity due to the decoy of different miRNAs [28][29][30]. (3) Guide lncRNAs can regulate gene expression through the recruitment and re-localization of ribonucleoprotein complexes at specific chromatin loci, such as
MEG3
Table 1) [18][31][32]. (4) Scaffold lncRNAs can act as central platforms upon the assembly of different ribonucleoprotein complexes, affecting their molecular components (
HOTAIR
Table 1), promoting gene repression [18][26].
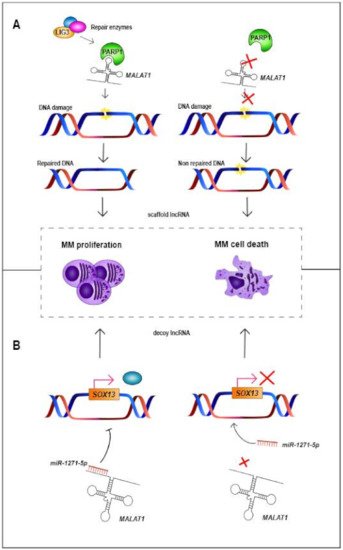
Figure 1.
MALAT1
A
MALAT1
MALAT1
MALAT1 and PARP1 does not occur, damaged DNA is not repaired, triggering MM cell death [33]. (
B
MALAT1
miR-1271-5p
MALAT1
miR-1271-5p
MALAT1
miR-1271-5p
Table 1.
| Gene | Location | Gene Type | Expression in MM | Molecular Mechanism | Molecular Interaction in MM | Biological Effect after lncRNA KD | Biological Effect after lncRNA UR | Prognosis in MM | References |
|---|
| ANRIL | 9p21.3 | Antisense | Up | Decoy | Binds to miR-34a, miR-125a, miR-186 and miR- 411–3p |
Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [34][35][33,34] |
| BM742401 | 18q11.2 | LincRNA | Down | NA | NA | NA | Decreases cell migration | Methylated lncRNA associated with worse OS | [36][35] |
| Circ_0000190 | 1q42.12 | Circular lncRNA | Down | NA | NA | NA | NA | High expression levels associated with better PFS and OS | [37][38][39][36,37,38] |
| CRNDE | 16q12.2 | LincRNA | Up | Decoy | Binds to miR-451 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse OS | [40][41][39,40] |
| DARS-AS1 | 2q21.3 | Antisense | Up under hypoxia | Decoy | Interacts with RBM39 and HIP-1α, suppressing mTOR pathway | Decreases cellular proliferation and increases apoptosis. Decreases tumorigenesis in vivo. Its upregulation reduces the sensitivity to bortezomib in vitro | NA | NA | [42][41] |
| ENSG00000249988 | 4p15.33 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS andbetter OS | [13] |
| ENSG00000254343 | 8q24.12 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS | [13] |
| FEZF1-AS1 | 7q31.32 | Antisense | Up | Decoy | Binds to miR-610 and regulates AKT3 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [43][42] |
| GAS5 | 1q25.1 | Processed transcript | Down | NA | NA | NA | Decreases cellular proliferation | NA | [44][43] |
| H19 | 11p15.5 | Processed transcript | Up | Decoy | Binds to miR-152-3p and miR-29b-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [45][46][47][44,45,46] |
| HOTAIR | 12q13.31 | Antisense | Up | NA | Activates NF-κB pathway | Decreases cellular proliferation, triggers cell cycle arrest and decreases chemoresistance to dexamethasone | NA | NA | [21][48][49][50][21,47,48,49] |
| HOXB-AS1 | 17q21.32 | Antisense | Up | Scaffold | Scaffold for ELAVL1. Interacts with FUT4-mediated Wnt/β-catenin pathway | Decreases cellular proliferation and increases apoptosis | NA | NA | [51][50] |
| IRAIN | 15q26.3 | Antisense | Down | Decoy | Binds to miR-125b and regulates IGF-1 signaling | NA | Increases apoptosis | NA | [52][53][51,52] |
| LINC00152 | 2p11.2 | LincRNA | Up | Decoy | Binds to miR-497 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumorigenesis in vivo | NA | High expression levels associated with worse OS | [54][53] |
| LINC00461 | 5q14.3 | LincRNA | Up | NA | NA | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [55][56][57][54,55,56] |
| LINC00515 | 21q21.3 | LincRNA | Up | Decoy | Binds to miR-140-5p | Increases apoptosis | NA | NA | [58][57] |
| LINC00665 | 19q13.12 | LincRNA | Up | Decoy | Binds to miR-214-3p | Decreases cellular proliferation and increases apoptosis | NA | NA | [59][58] |
| LINC01234 | 12q24.13 | LincRNA | Up | Decoy | Binds to miR-124-3p | Decreases cellular proliferation and increases apoptosis. Decreases cell proliferation and tumor growth in vivo | NA | High expression levels associated with worse OS | [60][59] |
| lnc-ANGPTL1-3 | 1q25.2 | Antisense | Up | Decoy | Binds to miR-30a-3p | Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [61][60] |
| lnc-TCF7 | 5q31.1 | NA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [62][61] |
| LUCAT1 | 5q14.3 | LincRNA | Up | NA | Activates the TGF-β signaling pathway | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with shorter five-year survival | [63][62] |
| MALAT1 | 11q13.1 | LincRNA | Up | Decoy and Scaffold | Binds to miR-1271-5p, miR-181a-5p and miR-509- 5p. Scaffold for PARP1 |
Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [64][65][66][67][68][69][70][33][63,64,65,66,67,68,69,70] |
| MEG3 | 14q32.2 | LincRNA | Down | Decoy | Binds to miR-181a | NA | Decreases cellular proliferation and increases apoptosis | High expression levels associated with better PFS and OS | [71][72][73][74][71,72,73,74] |
| MIAT | 22q12.1 | LincRNA | Up | Decoy | Binds to miR-29b | Sensitizes MM cells to bortezomib | NA | High expression levels associated with worse PFS and OS | [75][76][75,76] |
| NEAT1 | 11q13.1 | LincRNA | Up | Decoy | Binds to miR-214 and miR-125a | Decreases cellular proliferation | NA | High expression levels associated with worse PFS and OS | [77][78][79][80][77,78,79,80] |
| OIP5-AS1 | 15q15.1 | Processed transcript | Down | Decoy | Binds to miR-410 and miR-27a-3p | NA | Decreases cellular proliferation and increases apoptosis | NA | [81][82][81,82] |
| PCAT-1 | 8q24.21 | LincRNA | Up | Decoy | Binds to miR-129 | Increases apoptosis and sensitizes MM cells to bortezomib | NA | NA | [83][84][83,84] |
| PDIA3P | 1q21.1 | Pseudogene | Up | NA | NA | Decreases cellular proliferation. Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [85] |
| PDLIM1P4 | 3q12.1 | Pseudogene | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [13] |
| PRAL | 17p13.1 | NA | Down | Decoy | Binds to miR-210 | NA | Decreases cellular proliferation and increases apoptosis. Increases the anti-tumor effect of bortezomib | High expression levels associated with better PFS and OS | [86][87][86,87] |
| PVT1 | 8q24.21 | Processed transcript | Up | Decoy | Binds to miR-203a. It is inhibited by BRD4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [88][89][88,89] |
| SMILO | 1q42.2 | LincRNA | Up | NA | Regulates IFN pathway | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with better OS | [13] |
| SNHG16 | 17q25.1 | Processed transcript | Up | Decoy | Binds to miR-342-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [90] |
| SOX2OT | 3q26.3 | Sense overlapping | Up | Decoy | Binds to miR-144-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumor growth in vivo | NA | NA | [91] |
| ST3GAL6-AS1 | 3q12.1 | Antisense | Up | NA | NA | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [92][93][92,93] |
| TUG1 | 22q12.2 | Antisense | Up | Decoy | Binds to miR-29b-3p and targets HDAC4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [44][94][43,94] |
| UCA1 | 19p13.12 | Processed transcript | Up | Decoy | Binds to miR-1271-5p and miR-331-3p | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [95][96][95,96] |
| XLOC_013703 | 20p11.21 | NA | Down | NA | Involved in NF-κB signaling activation | NA | Decreases cellular proliferation and increases apoptosis | NA | [97] |
3. Role of lncRNAs in the Pathobiology of MM
Diverse studies have pointed to the importance of lncRNAs in different biological processes, such as immune response, cell differentiation, gene expression modulation and chromatin reorganization [98][99]. Intriguingly, their deregulation also contributes to the development of carcinogenesis, metastasis and even anti-cancer treatment resistance [64]. The deregulation of the expression of lncRNAs can thus impact on relevant pathways involved in the pathogenesis and/or progression of certain human tumors, including MM [44][71]. We have recently demonstrated that 82% of the transcriptome, including coding genes and all types of polyA+ and non-polyA lncRNAs, in plasma cells from MM correspond to lncRNAs, compared to 18% of coding genes [13].
HOTAIR
PDIA3P
LINC00461 in both HCC and lung cancer, all three of them being also upregulated in MM [48][49][50][55][56][57][100].
PRAL
GAS5 (growth arrest specific 5) in breast, prostate, renal cancer and MM [44][86][87][101][102]. However, there are lncRNAs that are deregulated in MM while they show the opposite direction of expression in other neoplasms. For instance,
MALAT1 is upregulated in MM, lung cancer, gallbladder cancer, colorectal carcinoma and HCC, whilst this lncRNA is downregulated in colorectal and glioma cancer [65][103][104].
NEAT1
Circ_0000190 is downregulated in MM and gastric cancer, whereas it displays overexpression in lung cancer [37][38][39]. These results highlight the cell- and tissue- specificity of lncRNAs, showing that their deregulation—and thus, their potential function—needs to be addressed in each tumor. For example,
MALAT1
NEAT1
MALAT1
miR-1271-5p
miR-181a-5p
miR-509-5p
miR-195
miR-206 and miR-363-3p in gallbladder cancer [64][66][67][68][69]. In the case of
NEAT1
miR-125a
miR-193a-3p in lung adenocarcinoma, among others [77][106]. Usually, the expression of lncRNAs and miRNAs is negatively correlated. Therefore, overexpression of one lncRNA could trigger the downregulation of miRNAs, whereas downregulation of one lncRNA could promote the overexpression of different miRNAs [34][77]. Likewise, there are other examples of lncRNAs which act as miRNA sponges in MM (
CRNDE
IRAIN
H19
UCA1
OIP5-AS1
TUG1
H19
miR-29b-3p
Table 1) [34][40][43][45][46][52][54][58][59][60][61][72][81][82][83][88][90][91][94][95][96]. These results highlight the relevance of the miRNA sponge function of lncRNAs in MM.
DARS-AS1
LINC00152
SOX2OT
LINC01234
miR-124-3p
GRB2
Table 1) [13][34][35][36][40][41][42][43][44][45][46][49][51][52][54][57][58][59][60][63][70][72][78][79][81][82][83][84][85][86][88][90][91][92][94][95][96][97][107].
