Alzheimer’s disease (AD) is the most prevalent form of dementia; however, the precise pathophysiology of the disease has not been elucidated yet. Thus, understanding the disease pathology, as well as identification and development of valid biomarkers, are necessary for identifying therapeutic targets and early diagnosis as well as for monitoring disease progression. Mass spectrometry (MS)-based proteomics studies on clinical specimens, in-vitro and in-vivo models of AD has greatly aided such efforts.
- Alzheimer
- mass spectrometry
- proteomics
- targeted proteomics
1. Introduction
- Introduction
Alzheimer’s disease (AD) is the most common progressive neurodegenerative disorder with memory loss, cognitive impairment, disorientation, and psychiatric symptoms. These symptoms render AD patients incapable of independently performing day-to-day activities. With increasing average life expectancy, it is becoming one of the greatest healthcare challenges of the 21st century [1]. The major hallmark of AD is extracellular deposition of amyloid-β (Aβ) as plaques in the brain and the formation of intracellular neurofibrillary tangles (NFTs) composed of the accumulation of hyperphosphorylated tau (p-tau) [2][3][2,3]. Despite advancements in research, definitive causes of the disease remain elusive with no effective treatments available yet [4]. As the understanding of AD pathogenesis is limited, it is difficult to develop novel therapies for AD. Therefore, further understanding of molecular mechanisms and cellular signalling pathways that are responsible for AD pathogenesis is needed for discovering new targets and evolving treatment strategies for AD. Mass-spectrometry-based proteomics will continue to play an important role in the future for discovering these events responsible for AD pathogenesis.
2. Proteomics Studies in AD Pathogenesis and Biomarker Discovery
- Proteomics Studies in AD Pathogenesis and Biomarker Discovery
Over the past few decades, several studies on both patient-derived biological samples as well as in vivo and in vitro Alzheimer’s disease models utilizing proteomics technologies have been carried out. These studies have contributed greatly toward understanding the disease pathogenesis as well as the identification of biomarkers (Figure 1).
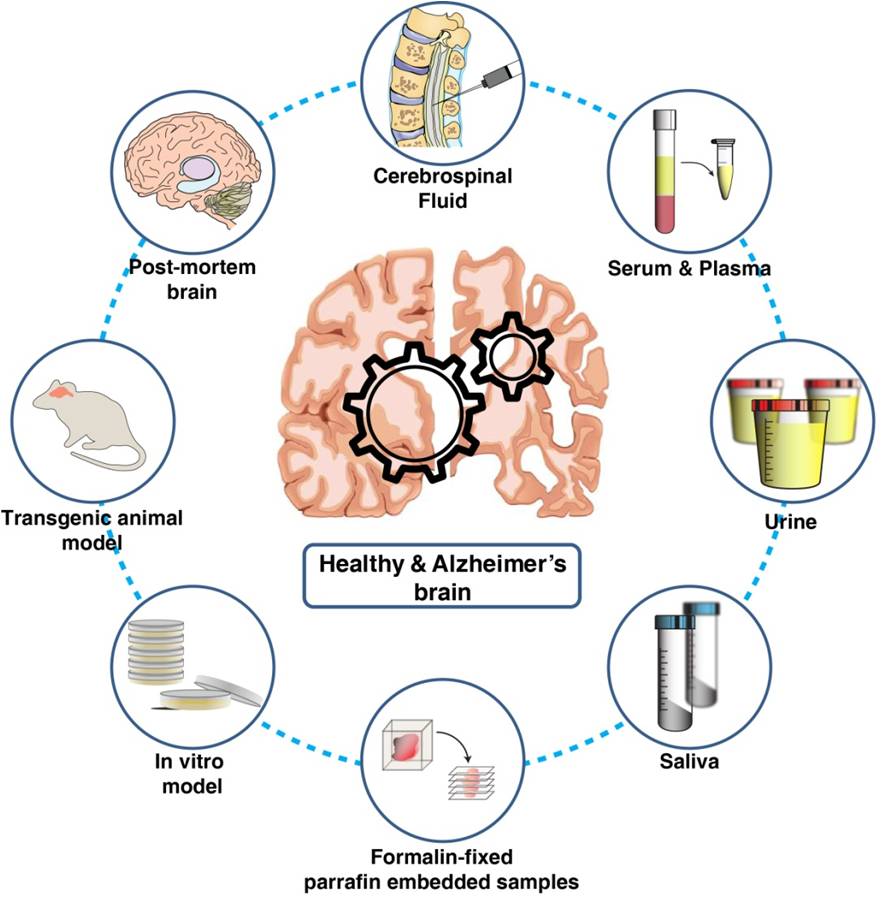
Figure 1. Biological materials used for understanding Alzheimer’s pathology and for the discovery and validation of candidate biomarkers using proteomics technologies.
2.1. Proteomics and AD Pathogenesis
The past decade has seen remarkable advances in high-resolution mass-spectrometry-based proteomics that have enabled precise analysis of thousands of proteins from brain tissue samples in a relatively shorter amount of time. Understanding the function of the brain and the related disease mechanisms requires comprehensive knowledge of the changes in the neuronal proteome and post-translational modifications (PTMs). Quantitative proteome analysis of the human hippocampus at different Braak stages has demonstrated alteration in components of the extracellular matrix and calcium-dependent signalling [5]. Multiple large-scale deep proteomics studies have demonstrated the involvement of diverse signalling networks including proteostasis, RNA homeostasis, neuroinflammation, lipid homeostasis, and myelin-axon interactions among others to be associated with Alzheimer’s pathology [6][7][8][9][10][11][12][6-12]. These studies show that a gamut of cellular protein networks and processes are associated with the pathology of this disease. Apart from brain tissues, protein aggregates such as extracellular amyloid-beta plaques and tau neurofibrillary tangles have also been extensively studied using proteomics techniques providing insights into their role in AD pathology and progression [13]. Deep proteome analysis of detergent-insoluble protein aggregates from brain tissue identified ~4000 proteins, including U1–70K and other U1 small nuclear ribonucleoprotein (U1 snRNP) spliceosome components in AD as well as mild cognitive impairment (MCI) samples [14]. Such insights underscore the need for further functional studies to probe the exact role of aggregate-associated proteins in AD.
Studies on human postmortem brain tissue have greatly improved our understanding of the disease, however; there are several limitations to use brain tissue for research purposes. Major limitations being the availability of tissue, quality of the tissue, genetic heterogeneity, pre-terminal medication or concomitant disease, cause of death, and postmortem interval. In-vivo animal model systems serve as a better alternative to postmortem brain tissue in understanding disease mechanisms as many of these issues related to consistency can be controlled. Multiple mouse models have been employed for studying AD-related neurodegeneration. Li et al. carried out proteome profiling of hippocampus of APPSw,Ind J20 mouse and identified alterations in mTOR signalling and β-spectrin, Rab3-mediated APP trafficking, and proteolysis as early molecular events associated with AD pathogenesis [15]. Alteration in synaptic functions associated with dendritic spine morphology, neurite outgrowth, long-term potentiation, CREB signalling, and cytoskeletal dynamics was revealed by systematic analysis of proteome, phosphoproteome, and sialylated N-linked glycosylation of APPswe/PS1ΔE9 mouse olfactory bulb, hippocampus, neocortex, and the brainstem [16].
Although the use of such animal models has been extremely useful translational clinical research is still heavily reliant on human tissue specimens. One of the other alternatives to fresh human tissue is a formalin-fixed and paraffin-embedded (FFPE) tissue sample. These FFPE tissues can be stored for the long term and also have a plethora of associated clinical data like survival time and therapy response. Drummond et al. isolated neuronal cells from temporal cortex FFPE samples with severe Alzheimer’s using the LCM-LC-MS/MS technique and identified more than 400 proteins of which 78% were of neuronal origin and about 200 proteins were associated with Alzheimer’s disease [17]. However, further challenges that are yet to be reliably addressed include limiting the effects of formaldehyde-induced protein modification on protein recovery as well as identification in shotgun proteomics [18]. Another relevant area that is still largely unexplored in the context of Alzheimer’s disease is the use of FFPE samples for the identification of changes in the post-translation modifications.
2.2. Proteomics and Fluid Biomarkers Discovery
The goal of the protein marker discovery is to detect a protein or panel of proteins that differentiate affected patients with a specific disease from healthy individuals. For biomarker analysis, it is preferred that it would be accessible through minimal invasion.
As cerebrospinal fluid (CSF) is in close proximity to brain tissue it has been extensively probed in search of biomarkers for neurological diseases including AD. There are several studies published on deep proteomic analysis of CSF using depletion of high-abundant proteins coupled with high-resolution mass spectrometry to increase coverage of low-abundant proteins [19][20][19,20]. Previously known AD markers including Tau and amyloid-beta peptides have been identified across multiple mass-spectrometry-based proteomics studies and have been successfully developed into targeted assays for quantitation [21][22][23][24][21-24]. Barthelemey et al. have studied tau phosphorylation in CSF, brain, and plasma and reported an association between site-specific changes in tau phosphorylation and evolution of stages of dominantly inherited Alzheimer’s disease [25][26][27][25-27]. Although CSF-biomarker studies have identified several novel protein markers for AD, these studies are largely constrained by limited sample size.
In neurodegenerative disease, the blood-brain barrier is disturbed which results in increased permeability, and greater exchange of proteins between blood and CSF [28]. Several discovery and targeted proteomics studies have been carried out for the identification of serum protein markers despite the enormous challenge of a large dynamic range of protein abundance that spans 12 orders of magnitude [29][30][31][32][33][29-33]. Zetterberg et al. has curated a timeline for recent biomarker studies aimed at the development of clinically implementable blood test for Alzheimer’s summarizing different technologies employed for such research [34]. Dey et. al. used extensive pre-fractionation-based deep proteomic profiling of undepleted human serum for the biomarker discovery for Alzheimer’s disease. They reported a panel of 30 significantly altered proteins related to mitochondrial function including downregulation of AK2 and PCK2 in Alzheimer’s samples [35]. These studies, although limited by sample size, demonstrate serum biomarkers as a viable option despite several challenges posed by being a complex matrix.
Apart from CSF and serum other body fluids such as urine, saliva, and ocular fluids have also been probed in search of candidate biomarkers. Compared to both blood and CSF these fluids are comparatively very easily accessible through noninvasive procedures. Increased levels of urinary Aβ42 have been demonstrated in AD samples and proposed as a marker for both AD diagnosis and monitoring [36]. Expression of tau mRNA and amyloid-β precursor protein (AβPP) has been reported in the salivary gland and human salivary epithelial cells, respectively [37][38][37,38]. Min Shi et al. have also reported the identification of salivary tau and p-tau as AD biomarkers by immunoprecipitation and mass-spectrometry-based identification [39]. Visual symptoms have been reported in many AD patients, and this has piqued an increased interest in discovering ocular biomarkers that might be related to the pathology [40][41][40,41]. Several proteomics studies have been carried analyzing the tear proteome to identify AD markers [42][43][42,43]. These studies demonstrate the feasibility of using these alternative fluids as targets for biomarker discovery. Such markers also have the potential to be used for screening asymptomatic subjects, dramatically increasing the window for therapeutic interventions.
3. Proteomics Technologies Employed for Understanding AD Pathogenesis and Biomarker Discovery
- Proteomics Technologies Employed for Understanding AD Pathogenesis and Biomarker Discovery
Proteomics technologies that are employed toward the identification of the AD-related changes in CSF, plasma, and tissue are broadly categorized as discovery or targeted proteomics (Figure 2). The discovery-based approach has been primarily used for the quantitation of changes in molecular signalling networks that are associated with AD pathology. While a targeted proteomics approach is being used for monitoring and quantitation of proteins that are already known to be associated with AD pathogenesis and can be used as a candidate marker.
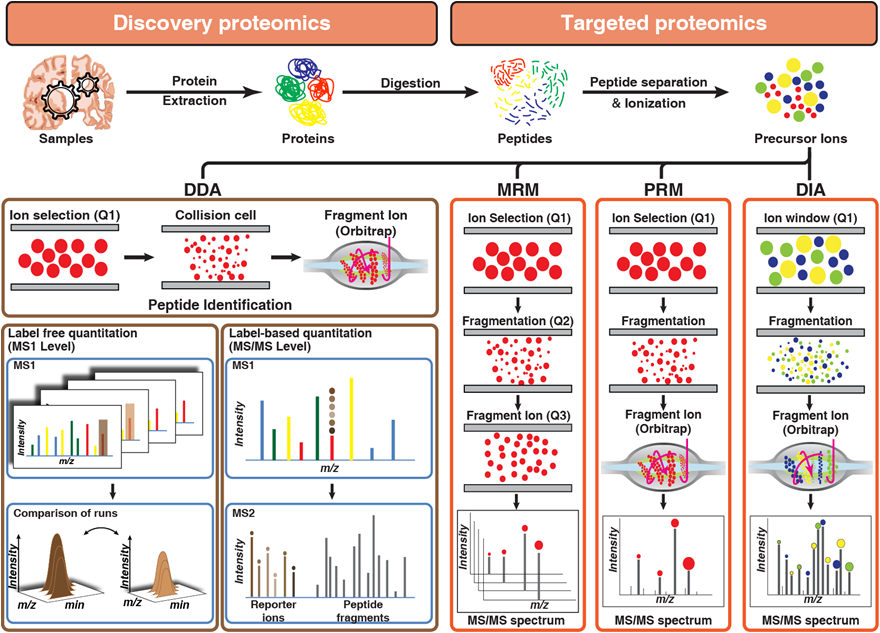
Figure 2. Overview of discovery and targeted proteomics technologies: Panel 1 describes label-free and label-based quantitation techniques used in discovery proteomics to identify differentially expressed proteins across samples by comparing either intensities and peak area of peptides (label-free quantitation) or reporter ion intensities (label-based quantitation) generated after fragmentation of labeled peptide precursors. Panel 2 describes targeted proteomics techniques used for monitoring peptides across samples by either sequential selection and monitoring of precursor-product ion pair (MRM) or simultaneous monitoring of all fragment ions of a selected precursor ion (PRM) or targeted extraction of fragment ion intensities from a mixed spectrum of fragment ions generated from multiple precursor ions selected by using a small window of m/z (DIA).
3.1. Discovery-Based Proteomics Analyses
Data-dependent acquisition or information-dependent acquisition has been the cornerstone of shotgun-discovery-based proteomics technology. In the data-dependent acquisition (DDA) the instrument cycles through a short high-resolution MS1 survey scan of the peptides followed by series of quick low-resolution MS2 scans for detection of the fragment ions of peptides selected based on the intensity. Label-free proteomic analysis approach has been used for quantitation of proteins for Alzheimer’s disease (AD) brains versus normally aged brains [44] and membrane-enriched proteome from postmortem human brain tissue in Alzheimer’s disease [45]. Several studies have carried out proteomics analysis for the discovery of biomarkers using either iTRAQ or TMT-based multiplexing approach. Sathe et. al. identified several known and novel CSF protein biomarker including MAPT, NPTX2, PKM, and YWHAG using TMT-based quantitative proteomics [19]. Adav et. al. used iTRAQ-based quantitative proteomics for identifying alterations in mitochondrial proteome of the human brain tissues of healthy and AD individuals.
Apart from the changes in cellular protein levels characterization of changes in protein post-translational modifications (PTMs) is needed for the understanding of the complex regulatory circuits that drive disease pathology. There are more than 200 types of PTMs that are known to control cellular functions. Domenico et al. identified significant alteration in neuronal processes such as formation, outgrowth, and guidance of neurites on quantitative phosphoproteomics analysis of the hippocampus of the Alzheimer’s disease subjects [46]. Kempf et al. carried out proteomics, phosphoproteomics, and N-glycosylation changes in the four different parts of the brain of the APP/PS1 Alzheimer’s mouse model identifying a brain-region specific response in CREB-mediated synaptic signalling [47]. Chou et al. have reviewed the effects of multiple post-translational modifications including phosphorylation, acetylation, SUMOylation, O-GlcNAcylation, and ubiquitination of tau on AD pathology [48]. Rayaprolu et al. describe how a network-based proteomics approach can be leveraged to improve our narrow understanding of AD biology beyond amyloid plaques and tau phosphorylation [49]. One of the major advantages of PTM proteomics studies is that the altered kinases identified by them will be also helpful as potential therapeutic targets in the future.
3.2. Targeted Proteomics Analyses
Once the discovery data are acquired, rigorous statistical methods are applied to identify significantly differential proteins that can be used as potential markers for the disease. To quantitate the abundance of these markers reproducibly, highly robust and reproducible methods are required. In general proteomics, workflow DDA and targeted proteomics are used in tandem or parallel. In targeted proteomics, multiple- or parallel-reaction monitoring (MRM or PRM) and data-independent analysis (DIA) are generally used (Figure 2).
In targeted proteomic analysis such as MRM or PRM precursor ions of selected peptides are filtered and fragmented and only desired fragment ions recorded. An increase in CSF concentrations of Alpha-, Beta-, and Gamma-synuclein has been demonstrated in Alzheimer’s patients using the MRM-proteomics approach [50]. Similarly, Wesseling et al. reported heterogeneity on tau phosphorylation sites across AD patients using FLEXItau, an MRM-based assay [51][52][51,52]. Using a label-free PRM approach Sathe et. al. demonstrated that NPTX2, in combination with PKM or YWHAG in CSF differentiate AD from control samples [19]. Because of their high specificity and multiplexing abilities, targeted proteomics assays have become a very valuable tool for assessing proteins for which commercial antibodies are not available.
Data-independent analysis (DIA) is a recent advancement and a rapidly growing technology in proteomics analysis [53]. In DIA experiments MS/MS data from all detectable precursor ions present in a biological sample are recorded generating a complete and permanent digital archive of the sample. Chang et al. used DIA for quantifying the synaptosomal proteome from the hippocampus and motor cortex tissue from Alzheimer’s subjects. They reported differential expression of 30 proteins including 17 novel proteins in AD [54]. Bader et al. characterized CSF proteome changes that accompany AD in three independent cohorts consistently quantifying more than 1000 proteins [55]. They reported 20 proteins to be differentially expressed in AD-CSF samples including tau, SOD1, PARK7, YKL-40, and novel biomarker candidates such as YWHAG, PKM, and ALDOC. Bader et al. provide a novel application of DIA proteomics strategies to drive a shift in the clinical proteome paradigm toward a population-wide discovery and validation of biomarker candidates that exonerate the individual-specific effects observed in small pilot studies.
4. Future Directions
- Future Directions
Recent developments in proteomics sample preparation methodologies along with advances in high-throughput mass spectrometry have improved the proteome coverage. Several quantitative proteomics methods including label-free, isobaric labelling based in the discovery platform and MRM, PRM, and DIA in the targeted platform have been employed for AD biomarker discovery and understanding AD pathogenesis. These advancements in technologies have rapidly generated several datasets on various biofluids, tissues, and animal models related to Alzheimer’s disease. Although these studies have dramatically improved our understanding of disease pathology, as the complexity of the disease is high, studies at a large scale in diverse populations will be required in the future for the identification of robust biomarkers. One of the other limitations in the identification of robust and reliable biomarker candidates is the current “triangular strategy” of discovering candidates from a smaller subset of samples and validation in a larger cohort. Where many of the identified candidates turn out to be specific to the discovery cohort and fail at validation stages in larger cohorts. Such limitations are now being addressed by a shift in the study design of clinical discovery proteomics with a “rectangular strategy”. Such strategies employ high proteome depth workflows for simultaneous discovery and validation on hundreds of samples in a population-wide setting. In the future, it is also necessary to have a stringent reporting system for study and patient selection criteria, clinical metadata, and parameter used for analysis. This information will be useful for comparison of the datasets as well as to build a knowledge base that will help to further our understanding of the disease and associated neurodegeneration.
References
- Scheltens, P.; Blennow, K.; Breteler, M.M.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer's disease. Lancet 2016, 388, 505-517, doi:10.1016/S0140-6736(15)01124-1.
- Glenner, G.G.; Wong, C.W. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun 1984, 120, 885-890.
- Lee, V.M.; Balin, B.J.; Otvos, L., Jr.; Trojanowski, J.Q. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science 1991, 251, 675-678.
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer's disease. Wiley Interdiscip Rev RNA 2018, 9, doi:10.1002/wrna.1463.
- Hondius, D.C.; van Nierop, P.; Li, K.W.; Hoozemans, J.J.; van der Schors, R.C.; van Haastert, E.S.; van der Vies, S.M.; Rozemuller, A.J.; Smit, A.B. Profiling the human hippocampal proteome at all pathologic stages of Alzheimer's disease. Alzheimers Dement 2016, 12, 654-668, doi:10.1016/j.jalz.2015.11.002.
- Zhang, Q.; Ma, C.; Gearing, M.; Wang, P.G.; Chin, L.S.; Li, L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer's disease. Acta Neuropathol Commun 2018, 6, 19, doi:10.1186/s40478-018-0524-2.
- Sathe, G.; Albert, M.; Darrow, J.; Saito, A.; Troncoso, J.; Pandey, A.; Moghekar, A. Quantitative proteomic analysis of the frontal cortex in Alzheimer's disease. J Neurochem 2020, 10.1111/jnc.15116, doi:10.1111/jnc.15116.
- Wang, H.; Dey, K.K.; Chen, P.C.; Li, Y.; Niu, M.; Cho, J.H.; Wang, X.; Bai, B.; Jiao, Y.; Chepyala, S.R., et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer's disease. Mol Neurodegener 2020, 15, 43, doi:10.1186/s13024-020-00384-6.
- Bai, B.; Wang, X.; Li, Y.; Chen, P.C.; Yu, K.; Dey, K.K.; Yarbro, J.M.; Han, X.; Lutz, B.M.; Rao, S., et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron 2020, 106, 700, doi:10.1016/j.neuron.2020.04.031.
- Wang, Z.; Yu, K.; Tan, H.; Wu, Z.; Cho, J.H.; Han, X.; Sun, H.; Beach, T.G.; Peng, J. 27-Plex Tandem Mass Tag Mass Spectrometry for Profiling Brain Proteome in Alzheimer's Disease. Anal Chem 2020, 92, 7162-7170, doi:10.1021/acs.analchem.0c00655.
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Ping, L.; Zhou, M.; Yin, L.; Higginbotham, L.A.; Guajardo, A.; White, B.; Troncoso, J.C., et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med 2020, 26, 769-780, doi:10.1038/s41591-020-0815-6.
- Higginbotham, L.; Ping, L.; Dammer, E.B.; Duong, D.M.; Zhou, M.; Gearing, M.; Hurst, C.; Glass, J.D.; Factor, S.A.; Johnson, E.C.B., et al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer's disease. Sci Adv 2020, 6, doi:10.1126/sciadv.aaz9360.
- Lutz, B.M.; Peng, J. Deep Profiling of the Aggregated Proteome in Alzheimer's Disease: From Pathology to Disease Mechanisms. Proteomes 2018, 6, doi:10.3390/proteomes6040046.
- Bai, B.; Hales, C.M.; Chen, P.C.; Gozal, Y.; Dammer, E.B.; Fritz, J.J.; Wang, X.; Xia, Q.; Duong, D.M.; Street, C., et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc Natl Acad Sci U S A 2013, 110, 16562-16567, doi:10.1073/pnas.1310249110.
- Li, N.; Hu, P.; Xu, T.; Chen, H.; Chen, X.; Hu, J.; Yang, X.; Shi, L.; Luo, J.H.; Xu, J. iTRAQ-based Proteomic Analysis of APPSw,Ind Mice Provides Insights into the Early Changes in Alzheimer's Disease. Curr Alzheimer Res 2017, 14, 1109-1122, doi:10.2174/1567205014666170719165745.
- Kempf, S.J.; Metaxas, A.; Ibanez-Vea, M.; Darvesh, S.; Finsen, B.; Larsen, M.R. An integrated proteomics approach shows synaptic plasticity changes in an APP/PS1 Alzheimer's mouse model. Oncotarget 2016, 7, 33627-33648, doi:10.18632/oncotarget.9092.
- Drummond, E.S.; Nayak, S.; Ueberheide, B.; Wisniewski, T. Proteomic analysis of neurons microdissected from formalin-fixed, paraffin-embedded Alzheimer's disease brain tissue. Sci Rep 2015, 5, 15456, doi:10.1038/srep15456.
- Giusti, L.; Angeloni, C.; Lucacchini, A. Update on proteomic studies of formalin-fixed paraffin-embedded tissues. Expert Rev Proteomics 2019, 16, 513-520, doi:10.1080/14789450.2019.1615452.
- Sathe, G.; Na, C.H.; Renuse, S.; Madugundu, A.K.; Albert, M.; Moghekar, A.; Pandey, A. Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer's Disease. Proteomics Clin Appl 2018, 10.1002/prca.201800105, e1800105, doi:10.1002/prca.201800105.
- Khoonsari, P.E.; Haggmark, A.; Lonnberg, M.; Mikus, M.; Kilander, L.; Lannfelt, L.; Bergquist, J.; Ingelsson, M.; Nilsson, P.; Kultima, K., et al. Analysis of the Cerebrospinal Fluid Proteome in Alzheimer's Disease. PLoS One 2016, 11, e0150672, doi:10.1371/journal.pone.0150672.
- Zhou, M.; Haque, R.U.; Dammer, E.B.; Duong, D.M.; Ping, L.; Johnson, E.C.B.; Lah, J.J.; Levey, A.I.; Seyfried, N.T. Targeted mass spectrometry to quantify brain-derived cerebrospinal fluid biomarkers in Alzheimer's disease. Clin Proteomics 2020, 17, 19, doi:10.1186/s12014-020-09285-8.
- Lin, P.P.; Chen, W.L.; Yuan, F.; Sheng, L.; Wu, Y.J.; Zhang, W.W.; Li, G.Q.; Xu, H.R.; Li, X.N. An UHPLC-MS/MS method for simultaneous quantification of human amyloid beta peptides Abeta1-38, Abeta1-40 and Abeta1-42 in cerebrospinal fluid using micro-elution solid phase extraction. J Chromatogr B Analyt Technol Biomed Life Sci 2017, 1070, 82-91, doi:10.1016/j.jchromb.2017.10.047.
- Barthelemy, N.R.; Fenaille, F.; Hirtz, C.; Sergeant, N.; Schraen-Maschke, S.; Vialaret, J.; Buee, L.; Gabelle, A.; Junot, C.; Lehmann, S., et al. Tau Protein Quantification in Human Cerebrospinal Fluid by Targeted Mass Spectrometry at High Sequence Coverage Provides Insights into Its Primary Structure Heterogeneity. J Proteome Res 2016, 15, 667-676, doi:10.1021/acs.jproteome.5b01001.
- Oe, T.; Ackermann, B.L.; Inoue, K.; Berna, M.J.; Garner, C.O.; Gelfanova, V.; Dean, R.A.; Siemers, E.R.; Holtzman, D.M.; Farlow, M.R., et al. Quantitative analysis of amyloid beta peptides in cerebrospinal fluid of Alzheimer's disease patients by immunoaffinity purification and stable isotope dilution liquid chromatography/negative electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 2006, 20, 3723-3735, doi:10.1002/rcm.2787.
- Barthelemy, N.R.; Mallipeddi, N.; Moiseyev, P.; Sato, C.; Bateman, R.J. Tau Phosphorylation Rates Measured by Mass Spectrometry Differ in the Intracellular Brain vs. Extracellular Cerebrospinal Fluid Compartments and Are Differentially Affected by Alzheimer's Disease. Front Aging Neurosci 2019, 11, 121, doi:10.3389/fnagi.2019.00121.
- Horie, K.; Barthelemy, N.R.; Mallipeddi, N.; Li, Y.; Franklin, E.E.; Perrin, R.J.; Bateman, R.J.; Sato, C. Regional correlation of biochemical measures of amyloid and tau phosphorylation in the brain. Acta Neuropathol Commun 2020, 8, 149, doi:10.1186/s40478-020-01019-z.
- Barthelemy, N.R.; Li, Y.; Joseph-Mathurin, N.; Gordon, B.A.; Hassenstab, J.; Benzinger, T.L.S.; Buckles, V.; Fagan, A.M.; Perrin, R.J.; Goate, A.M., et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer's disease. Nat Med 2020, 26, 398-407, doi:10.1038/s41591-020-0781-z.
- Montagne, A.; Barnes, S.R.; Sweeney, M.D.; Halliday, M.R.; Sagare, A.P.; Zhao, Z.; Toga, A.W.; Jacobs, R.E.; Liu, C.Y.; Amezcua, L., et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015, 85, 296-302, doi:10.1016/j.neuron.2014.12.032.
- Kitamura, Y.; Usami, R.; Ichihara, S.; Kida, H.; Satoh, M.; Tomimoto, H.; Murata, M.; Oikawa, S. Plasma protein profiling for potential biomarkers in the early diagnosis of Alzheimer's disease. Neurol Res 2017, 39, 231-238, doi:10.1080/01616412.2017.1281195.
- Shen, L.; Liao, L.; Chen, C.; Guo, Y.; Song, D.; Wang, Y.; Chen, Y.; Zhang, K.; Ying, M.; Li, S., et al. Proteomics Analysis of Blood Serums from Alzheimer's Disease Patients Using iTRAQ Labeling Technology. J Alzheimers Dis 2017, 56, 361-378, doi:10.3233/JAD-160913.
- Hye, A.; Lynham, S.; Thambisetty, M.; Causevic, M.; Campbell, J.; Byers, H.L.; Hooper, C.; Rijsdijk, F.; Tabrizi, S.J.; Banner, S., et al. Proteome-based plasma biomarkers for Alzheimer's disease. Brain 2006, 129, 3042-3050, doi:10.1093/brain/awl279.
- Han, S.H.; Kim, J.S.; Lee, Y.; Choi, H.; Kim, J.W.; Na, D.L.; Yang, E.G.; Yu, M.H.; Hwang, D.; Lee, C., et al. Both targeted mass spectrometry and flow sorting analysis methods detected the decreased serum apolipoprotein E level in Alzheimer's disease patients. Mol Cell Proteomics 2014, 13, 407-419, doi:10.1074/mcp.M113.028639.
- Song, F.; Poljak, A.; Smythe, G.A.; Sachdev, P. Plasma biomarkers for mild cognitive impairment and Alzheimer's disease. Brain Res Rev 2009, 61, 69-80, doi:10.1016/j.brainresrev.2009.05.003.
- Zetterberg, H.; Blennow, K. Moving fluid biomarkers for Alzheimer's disease from research tools to routine clinical diagnostics. Mol Neurodegener 2021, 16, 10, doi:10.1186/s13024-021-00430-x.
- Dey, K.K.; Wang, H.; Niu, M.; Bai, B.; Wang, X.; Li, Y.; Cho, J.H.; Tan, H.; Mishra, A.; High, A.A., et al. Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer's disease. Clin Proteomics 2019, 16, 16, doi:10.1186/s12014-019-9237-1.
- Takata, M.; Nakashima, M.; Takehara, T.; Baba, H.; Machida, K.; Akitake, Y.; Ono, K.; Hosokawa, M.; Takahashi, M. Detection of amyloid beta protein in the urine of Alzheimer's disease patients and healthy individuals. Neurosci Lett 2008, 435, 126-130, doi:10.1016/j.neulet.2008.02.019.
- Oh, Y.S.; Turner, R.J. Effect of gamma-secretase inhibitors on muscarinic receptor-mediated calcium signaling in human salivary epithelial cells. Am J Physiol Cell Physiol 2006, 291, C76-82, doi:10.1152/ajpcell.00508.2005.
- Conrad, C.; Vianna, C.; Freeman, M.; Davies, P. A polymorphic gene nested within an intron of the tau gene: implications for Alzheimer's disease. Proc Natl Acad Sci U S A 2002, 99, 7751-7756, doi:10.1073/pnas.112194599.
- Shi, M.; Sui, Y.T.; Peskind, E.R.; Li, G.; Hwang, H.; Devic, I.; Ginghina, C.; Edgar, J.S.; Pan, C.; Goodlett, D.R., et al. Salivary tau species are potential biomarkers of Alzheimer's disease. J Alzheimers Dis 2011, 27, 299-305, doi:10.3233/JAD-2011-110731.
- Cronin-Golomb, A.; Sugiura, R.; Corkin, S.; Growdon, J.H. Incomplete achromatopsia in Alzheimer's disease. Neurobiol Aging 1993, 14, 471-477, doi:10.1016/0197-4580(93)90105-k.
- Schlotterer, G.; Moscovitch, M.; Crapper-McLachlan, D. Visual processing deficits as assessed by spatial frequency contrast sensitivity and backward masking in normal ageing and Alzheimer's disease. Brain 1984, 107 ( Pt 1), 309-325, doi:10.1093/brain/107.1.309.
- Kenny, A.; Jimenez-Mateos, E.M.; Zea-Sevilla, M.A.; Rabano, A.; Gili-Manzanaro, P.; Prehn, J.H.M.; Henshall, D.C.; Avila, J.; Engel, T.; Hernandez, F. Proteins and microRNAs are differentially expressed in tear fluid from patients with Alzheimer's disease. Sci Rep 2019, 9, 15437, doi:10.1038/s41598-019-51837-y.
- Kallo, G.; Emri, M.; Varga, Z.; Ujhelyi, B.; Tozser, J.; Csutak, A.; Csosz, E. Changes in the Chemical Barrier Composition of Tears in Alzheimer's Disease Reveal Potential Tear Diagnostic Biomarkers. PLoS One 2016, 11, e0158000, doi:10.1371/journal.pone.0158000.
- Andreev, V.P.; Petyuk, V.A.; Brewer, H.M.; Karpievitch, Y.V.; Xie, F.; Clarke, J.; Camp, D.; Smith, R.D.; Lieberman, A.P.; Albin, R.L., et al. Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains. J Proteome Res 2012, 11, 3053-3067, doi:10.1021/pr3001546.
- Donovan, L.E.; Higginbotham, L.; Dammer, E.B.; Gearing, M.; Rees, H.D.; Xia, Q.; Duong, D.M.; Seyfried, N.T.; Lah, J.J.; Levey, A.I. Analysis of a membrane-enriched proteome from postmortem human brain tissue in Alzheimer's disease. Proteomics Clin Appl 2012, 6, 201-211, doi:10.1002/prca.201100068.
- Di Domenico, F.; Sultana, R.; Barone, E.; Perluigi, M.; Cini, C.; Mancuso, C.; Cai, J.; Pierce, W.M.; Butterfield, D.A. Quantitative proteomics analysis of phosphorylated proteins in the hippocampus of Alzheimer's disease subjects. J Proteomics 2011, 74, 1091-1103, doi:10.1016/j.jprot.2011.03.033.
- Wang, F.; Blanchard, A.P.; Elisma, F.; Granger, M.; Xu, H.; Bennett, S.A.; Figeys, D.; Zou, H. Phosphoproteome analysis of an early onset mouse model (TgCRND8) of Alzheimer's disease reveals temporal changes in neuronal and glia signaling pathways. Proteomics 2013, 13, 1292-1305, doi:10.1002/pmic.201200415.
- Chu, D.; Liu, F. Pathological Changes of Tau Related to Alzheimer's Disease. ACS Chem Neurosci 2019, 10, 931-944, doi:10.1021/acschemneuro.8b00457.
- Rayaprolu, S.; Higginbotham, L.; Bagchi, P.; Watson, C.M.; Zhang, T.; Levey, A.I.; Rangaraju, S.; Seyfried, N.T. Systems-based proteomics to resolve the biology of Alzheimer's disease beyond amyloid and tau. Neuropsychopharmacology 2021, 46, 98-115, doi:10.1038/s41386-020-00840-3.
- Oeckl, P.; Metzger, F.; Nagl, M.; von Arnim, C.A.; Halbgebauer, S.; Steinacker, P.; Ludolph, A.C.; Otto, M. Alpha-, Beta-, and Gamma-synuclein Quantification in Cerebrospinal Fluid by Multiple Reaction Monitoring Reveals Increased Concentrations in Alzheimer's and Creutzfeldt-Jakob Disease but No Alteration in Synucleinopathies. Mol Cell Proteomics 2016, 15, 3126-3138, doi:10.1074/mcp.M116.059915.
- Mair, W.; Muntel, J.; Tepper, K.; Tang, S.; Biernat, J.; Seeley, W.W.; Kosik, K.S.; Mandelkow, E.; Steen, H.; Steen, J.A. FLEXITau: Quantifying Post-translational Modifications of Tau Protein in Vitro and in Human Disease. Anal Chem 2016, 88, 3704-3714, doi:10.1021/acs.analchem.5b04509.
- Wesseling, H.; Mair, W.; Kumar, M.; Schlaffner, C.N.; Tang, S.; Beerepoot, P.; Fatou, B.; Guise, A.J.; Cheng, L.; Takeda, S., et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer's Disease. Cell 2020, 183, 1699-1713 e1613, doi:10.1016/j.cell.2020.10.029.
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012, 11, O111 016717, doi:10.1074/mcp.O111.016717.
- Chang, R.Y.; Etheridge, N.; Nouwens, A.S.; Dodd, P.R. SWATH analysis of the synaptic proteome in Alzheimer's disease. Neurochem Int 2015, 87, 1-12, doi:10.1016/j.neuint.2015.04.004.
- Bader, J.M.; Geyer, P.E.; Muller, J.B.; Strauss, M.T.; Koch, M.; Leypoldt, F.; Koertvelyessy, P.; Bittner, D.; Schipke, C.G.; Incesoy, E.I., et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer's disease. Mol Syst Biol 2020, 16, e9356, doi:10.15252/msb.20199356.
