Metformin is the first-line pharmacotherapy for treating type 2 diabetes mellitus (T2DM); however, its mechanism of modulating glucose metabolism is elusive. Recent advances have identified the gut as a potential target of metformin. As patients with metabolic disorders exhibit dysbiosis, the gut microbiome has garnered interest as a potential target for metabolic disease. Henceforth, studies have focused on unraveling the relationship of metabolic disorders with the human gut microbiome.
- gut microbiome
- type 2 diabetes mellitus
- metformin
- dysbiosis
1. Introduction
Type 2 diabetes mellitus (T2DM) is one of the most common chronic metabolic disorders and is characterized by hyperglycemia resulting from the combination of insulin resistance and inadequate insulin secretion
. The number of people with T2DM has drastically increased over the past several decades
[1]
. Metformin, a biguanide class drug, is recommended by the American Diabetes Association and European Association for the Study of Diabetes as a first-line medicine for the treatment of T2DM
[2]
. Metformin is a derivative of phenformin and buformin from galegine in
Galega officinalis
, traditionally used to decrease blood sugar and relieve the symptoms of diabetes (
)
. Among the three biguanides, phenformin and buformin were withdrawn from the market due to the high frequency of lactic acidosis in the 1970s. However, metformin showed superior safety and better efficacy in the treatment of T2DM
. These advantages for clinical use have resulted in metformin being widely used for more than 60 years
[5]
. Metformin does not target a specific pathway or disease mechanism
[4]
; therefore, studies have aimed to reveal the mechanism of action of metformin related to the treatment of cancer and cardiovascular diseases
. Metformin exhibits the peak plasma concentrations in 3 h with C
max
1.0–1.6 mg/L for dose of 500 mg and approximately 55% of bioavailability (
[6]
and refences therein). After absorption, metformin is distributed in the liver, kidneys, adrenal glands, and pancreas at about seven-fold higher concentration than that of the serum
. Based on the evidence suggesting a higher accumulation of metformin in the liver as well as another report by Rena et al.
[9]
, the liver is a potential target organ of metformin
. Several studies have suggested that metformin suppresses the hepatic gluconeogenesis resulting from glucose tolerance modulation mediated by the adenosine monophosphate-activated protein kinase (AMPK) activity
. Recent evidence from three studies suggests that the gut is a major target of metformin action and not the liver. First, metformin when administered intravenously, instead of orally, demonstrated no glucose-lowering effects
. Further, the jejunum tissue was found to exhibit a metformin concentration of up to 2000 μmol/kg of tissue, which was 30–300 times higher than the plasma concentrations
. The jejunum biopsy under pre-dose and post-dose of metformin demonstrated the gastrointestinal tract as a prominent target of metformin
[18]
. Second, the organic cation transporter (OCT) 1, expressed in the membrane of enterocytes, might be possibly involved in the absorption of metformin from the intestinal lumen
. According to Dujic et al.
[22]
, a reduced function of OCT1 might increase the intestinal metformin concentration and the risk of gastrointestinal intolerance in the metformin-treated patients. Finally, the gut-restricted glucose-lowering effect of metformin was observed for intermediate-release metformin, extended-release metformin, and delayed-release metformin, and the same dose of metformin was more effective through those dosage forms than extended-release form
[23]
. Although various putative mechanisms of glucose homeostasis modulation in the gut by metformin have been proposed, more studies are needed to establish these hypotheses.
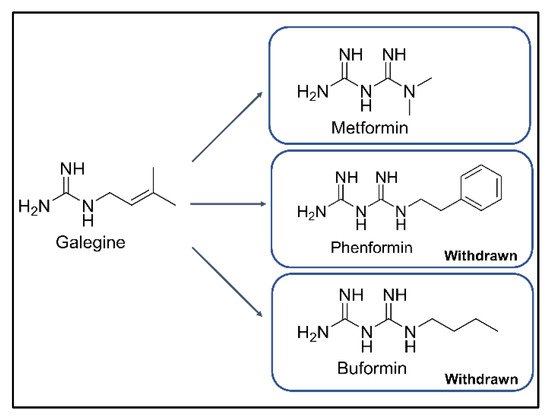
Figure 1. Chemical structures of galegine, metformin, phenformin, and buformin.
Microbiome in the human body assist in the expansion of host genomes, by facilitating the host’s metabolism and physiology
. Over the last few decades, the development of sequencing technologies and drastic progress in population-scale studies have revealed the host and microbiome relationship. Large-scale research projects on the microbiome have been actively conducted, such as the Human Microbiome Project (HMP) consortium funded by the United States National Institutes of Health (NIH) and the Metagenomics of the Human Intestinal Tract (MetaHIT) consortium funded by the European Commission
[24]
. A microbiome study demonstrated that the human gut microbiome abundance correlates with metabolic markers, such as adiposity, insulin resistance, and dyslipidemia
[26]
. Furthermore, gut dysbiosis has also been observed in T2DM patients
[27][28][29][30][31][32][33][34][35]
. Based on the hypothesis that metformin targets the human gastrointestinal tract, the gut microbiome has attracted attention as a key factor in the treatment of T2DM
. Thus, this review focused on the various studies related to the gut microbiome and its association with the anti-diabetic effects of metformin.
2. Gut Microbiome and T2DM
Over the past decade, several studies have demonstrated that patients with T2DM, obesity, or inflammatory bowel diseases often show dysbiosis in the gut microbiota
. The report by Larsen et al.
[30]
differentiated the composition of the gut microbiota in the T2DM patients from that in the non-diabetic adults (
), and other studies have demonstrated dysbiosis in T2DM patients under different conditions, such as subject’s race and co-administration with other drugs. According to Larsen et al.
[30]
, at the phylum level, the abundance of Firmicutes in T2DM patients was lower than that in the control group, and Bacteroidetes and Proteobacteria were more abundant than in the control group. The tendency of abundance at the phylum level was similar among the other clinical trials
. Furthermore, at the genus level,
Roseburia
, a butyrate-producing bacterium, was less abundant in the T2DM patients
. These results were in line with the other studies showing an increase in the abundance of
Roseburia
and insulin sensitivity after intestinal microbiota transplantation from lean donors to recipients with metabolic syndrome
[46]
. In addition, the abundance of
Lactobacillus
spp. was higher in T2DM patients than in the control groups
. The abundance of
Lactobacillus
spp. was positively correlated with blood glucose levels in the two clinical trials
and these results were consistent with those evident in a mice study
[47]
. The positive correlation between
Lactobacillus
spp. and the glucose levels might be due to the immunomodulatory role of
Lactobacillus
spp.
[48]
. Similarly, dysbiosis in T2DM patients might be due to the interaction of the gut microbiota with the host immune system, which was supported by several animal studies. In particular, the gut microbiota, which communicates with the host through pattern recognition receptors, such as toll-like receptors (TLRs), contributes to the development of insulin resistance with increased plasma LPS concentration
. According to Larsen et al.
[30]
, the abundance of Gram-negative bacteria, which can stimulate the immune system like TLRs, was increased in T2DM patients. The role of TLRs in insulin resistance has been established through various studies. The TLR-5 deficient mice became obese and exhibited a metabolic syndrome. Further, when the gut microbiome from the TLR-5 deficient mice was transplanted to the germ-free mice, the germ-free mice showed a similar phenomenon as the TLR-5 mice
[51]
. In addition, Song et al.
[52]
reported that TLR-4 activation is associated with insulin resistance in adipocytes. Previously cited clinical studies have identified SCFA-producing bacteria as the key for dysbiosis in T2DM patients in response to the immune responses
. The gut microbiota has been considered as one of the factors affecting T2DM; thus, the gut microbiota might be considered a potential target for the treatment of T2DM. Several studies have demonstrated the positive effects of probiotics for the treatment of T2DM, such as the decrease in systemic LPS levels and improvement in insulin resistance
.
Table 1. Alteration of the gut microbiota biochemical properties in the T2DM patients compared to the healthy subjects or alteration in the metformin treatment compared non-treatment T2DM patients or healthy subjects. ↑ (increase), ↓ (decrease), – (no alteration), NA (not applicable), Ref * (reference number).
| Ref * | Population | Study Design | Gut Microbiota | Biochemical Alterations |
|---|---|---|---|---|
| [27] | Chinese | T2DM patients (n = 170) |
versus Healthy subjects (n = 174) Family: Lachnospiraceae ↑, Erysipelotrichaceae ↓ Genus: Alistipes ↑, Clostridium ↑, Eubacterium ↓, Faecalibacterium ↓, Subdoligranulum ↑, Parabacteroides ↑ Species: Akkermansia muciniphila ↑, Bacteroides intestinalis ↑, Clostridium bolteae ↑, Clostridium hatheway ↑, Clostridium ramosum ↑, Clostridium symbiosum ↑, Eggerthella lenta ↑, Escherichia coli ↑, Eubacterium rectale ↓, Faecalibacterium prausnitzii ↓, Haemophilus parainfluenzae ↓, Roseburia intestinalis ↓, Roseburia inulinivorans ↓ |
NA |
| [30] | Danish | T2DM patients (n = 18) |
versus Healthy subjects (n = 18) α-diversity (Chao 1): – Phylum: Bacteroidetes ↑, Firmicutes ↓, Proteobacteria ↑ Class: Bacilli ↑, Bacteroidetes ↑, Betaproteobacteria ↑, Clostridia ↓ Genus: Akkermansia ↑, Alistipes ↑, Bacteroides ↓, Bifidobacterium ↓, Bilophila ↑, Catenibacterium ↓, Dialister ↑, Dorea ↑, Erysipelotrichaceae IS ↑, Faecalibacterium ↓, Lachnospiraceae IS ↓, Lactobacillus ↑, Parabacteroides ↑, Prevotella ↑, Roseburia ↓, Ruminococcus ↓, Sporobacter ↑, Subdoligranulum ↓, Succinivibrio ↑, Sutterella ↑ Species: Dorea longicatena ↓ |
NA |
| [32] | Chinese | T2DM patients (n = 13) |
versus Healthy subjects (n = 44) α-diversity (Chao 1, Shannon index): ↓ Class: Clostridia ↑, Clostridiales ↑ Family: Lachnospiraceae ↑ Genus: Abiotrophia ↑, Bacteroides ↓, Collinsella ↑, Dorea ↑, Eubacterium ↑, Haemophilus ↓, Megamonas ↓, Peptostreptococcus ↑, Prevotella ↑, Roseburia ↓, Ruminococcus ↑, Sporobacter ↑, Subdoligranulum ↑ |
NA |
| [34] | Pakistani | Obese-T2DM patients (n = 40) |
versus Healthy subjects (n = 20) α-diversity (Shannon index): ↓ Phylum: Bacteroidetes ↓, Elusimicrobia ↓, Firmicutes ↓, Proteobacteria ↓, Verrucomicrobioa ↓ Class: Bacilli ↓, Bacteroidia ↓, Clostridia ↑, Coriobacteriia ↑, Deltaproteobacteria ↓, Elusimicrobia ↓, Gammaproteobacteria ↓, Negativicutes ↑, Genus: Allisonella ↑, Bacillus ↓, Christensenellaceae_R_7 ↑, Dialister ↑, Escherichia_Shigella ↓, Eubacterium coprostanoligenes groups ↑, Lactobacillus ↑, Prevotella_9 ↓, Ruminococcus_2 ↓, Subdoligranulum ↑ |
NA |
| Metformin Treatment Effects in T2DM Patients | ||||
| [28] | Japanese | T2DM patients (n = 50) |
versus normal subjects (n = 50) Genus: Atopobium cluster ↓, Lactobacillus ↑, Prevotella ↓ Species: Clostridium coccoides ↓, Lactobacillus plantarum ↑, Lactobacillus reuteri ↑ |
Fecal organic acids ↓ Acetic acid ↓ Propionic acid ↓ Fecal isovaleric acid ↑ CRP ↑, IL-6 ↑ |
| Metformin treated T2DM (n = 17) | versus non treated T2DM (n = 33) Family: Enterobacteriaceae ↑ Genus: Staphylococcus ↑ Species: Clostridium coccoides ↓, Lactobacillus plantarum ↑, Lactobacillus reuteri ↑ |
NA | ||
| [29] | European old woman |
T2DM patients (n = 53) |
versus normal glucose tolerance (n = 43) Class: Clostridiales ↓ Family: Coriobacteriaceae ↓ Genus: Alistipes ↓, Clostridium ↓, Roseburia ↓, Species: Bacteroides intestinalis ↓, Eubacterium eligens ↓, Lactobacillus gasseri ↑, Streptococcus mutans ↑ |
C-peptide ↑ |
| Metformin treated T2DM (n = 20) |
versus non treated T2DM (n = 33) Genus: Clostridium ↓, Escherichia ↑, Eubacterium ↓, Klebsiella ↑, Salmonella ↑, Shigella ↑ Species: Escherichia coli ↑ |
NA | ||
| [31] | Danish | T2DM patients (n = 75) |
versus normal subjects (n = 277) Family: bp Clostridiales ↓, Peptostreptococcaceae ↓ Genus: Akkermansia ↓, Acidaminococcus ↑, Bilophila ↑, Collinsella ↑, Coprococcus ↓, Escherichia ↑, Holdemania ↑, Lactobacillus ↑, Parabacteroides ↑, Roseburia ↓, Veillonella ↓ |
NA |
| Metformin treated T2DM (n = 58) |
versus non treated T2DM (n = 17) Family: Peptostreptococcaceae ↓ Genus: Akkermansia ↑, Bilophila ↓, Escherichia ↑, Holdemania ↑, Roseburia ↑, Veillonella ↓ |
NA | ||
| Swedish female |
T2DM patients (n = 53) |
versus normal subjects (n = 92) Family: Peptostreptococcaceae ↓ Genus: Lactobacillus ↑ |
NA | |
| Metformin treated T2DM (n = 20) |
versus non treated T2DM (n = 33) Family: bp Clostridiales ↓, Peptostreptococcaceae ↓ Genus: Bilophila ↑, Escherichia ↑, Holdemania ↓, Lactobacillus ↑, Roseburia ↓, Veillonella ↓ |
NA | ||
| Chinese | T2DM patients (n = 71) |
versus normal subjects (n = 185) Family: bp Clostridiales ↓, Peptostreptococcaceae ↓, Genus: Acidaminococcus ↑, Bilophila ↑, Collinsella ↑, Coprococcus ↓, Escherichia ↑, Haemophilus ↓, Holdemania ↑, Lactobacillus ↑, Oscillibacter ↑, Roseburia ↓, Veillonella ↓ |
NA | |
| Metformin treated T2DM (n = 15) | versus non treated T2DM (n = 56) Family: bp Clostridiales ↑, Peptostreptococcaceae ↓ Genus: Bilophila ↑, Collinsella ↑, Escherichia ↓, Holdemania ↑, Parabacteroides ↑, Roseburia ↑, Subdoligranulum ↑, Veillonella ↓ |
NA | ||
| [33] | Chinese | T2DM patients (n = 26) |
versus normal subjects (n = 50) α-diversity (Shannon index): ↓ Phylum: Firmicutes ↓ Class: Fusobacteriia ↑ Family: Enterobacteriaceae ↓, Erysipelotrichaceae ↑, Erysipelotrichaceae ↑, Porphyromonadaceae ↑ Genus: Faecalibacterium ↓, Fusobacterium ↑, Lactobacillus ↑, Ruminococcus ↓ |
NA |
| Metformin treated T2DM (n = 51) |
versus non treated T2DM (n = 26) α-diversity (Shannon index): – Phylum: Actinobacteria ↓ Family: Enterobacteriaceae ↓, Spirochaetaceae ↑, Turicibacteraceae ↑ Genus: Fusobacterium ↑, Turicibacter ↑ |
NA | ||
| [55] | British | On metformin T2DM (visit 1 and 4, n = 12) |
versus off metformin T2DM (visit 2 and 3, n = 12) Genus: SMB53 ↓, Adlercreutzia ↓, Eubacterium ↑ |
Serum bile acids ↓ Fecal bile acids ↑ GLP-1 ↑ |
| [56] | Spanish | Metformin treated T2DM for 4 months (n = 22) | versus before metformin treatment in T2DM (n = 22) Phylum: Proteobacteria ↑, Firmicutes ↑ Genus: Actinetobacter ↑, Alkaliphilus ↓, Citrobacter ↑, Cronobacter ↑, Dermcoccus ↑, Desulfurispirillum ↑, Dickeya ↑, Edwardsiella ↑, Enterobacter ↑, Erwinia ↑, Escherichia ↑, Holdemania ↓, Intestinibacter ↓, Klebsiella ↓, Methylobaciilus ↑, Pantoea ↑, Pectobacterium ↑, Photorhabdus ↑, Providencia ↑, Pseudomonas ↑, Rahnella ↑, Rheinheimera ↑, Salmonella ↑, Subdoligranulum ↓, Xanthomonas ↑, Xenohabdus ↑, Yersinia ↑ Species: Akkermansia muciniphila ↑, Bifidobacterium adolescentis ↑ |
Fecal propionate, butyrate, lactate and succinate ↑ Plasma bile acids ↑ |
| [57] | Colombian | T2DM patients (n = 28) |
versus normal subjects (n = 84) Genus: Enterococcus casseliflavus ↓, Clostridiaceae 02d06 ↑, Prevotella ↑ |
NA |
| Metformin treated T2DM (n = 14) |
versus non treated T2DM (n = 14) Genus: Bacnesiellaceae ↓, Butyrivibrio ↑, Clostridiaceae 02d06 ↓, Megasphaera ↑, Oscillospira ↓, Prevotella ↑ |
NA | ||
| [58] | Scandinavian | Metformin treated T2DM (n = 23) |
versus non treated T2DM (n = 7) Family: Enterobacteriaceae ↑, Genus: Bacnesiellaceae ↓, Butyrivibrio ↑, Clostridiaceae 02d06 ↓, Megasphaera ↑, Oscillospira ↓, Prevotella ↑ |
SCFA concentration – |
| [59] | Chinese | Metformin treated for 3 days in T2DM (n = 22) | versus before metformin treatment in T2DM (n = 22) Genus: Bacteroides ↓ Species: Bacteroides fragilis ↓, Bacteroides finegoldii ↓, Bacteroides thetaiotaomicron ↓, Bacteroides uniformis ↓, Bacteroides ovatus ↓, Bacteroides intestinalis ↓, Bacteroides stercoris ↓, Bacteroides eggerthii ↓, Bacteroides fluxus ↓, Bacteroides caccae ↓, Bacteroides dorei ↓ |
GUDCA, Tauroursodeoxycholic acid, Conjugated Secondary bile acids ↑ Total bile acids – |
| Metformin Treatment Effects in Healthy Subjects | ||||
| [60] | Caucasian | Metformin treated for 7 days in healthy subjects (n = 18) |
versus before metformin treatment in healthy subjects (n = 18) α-diversity (Shannon index): ↓ Class: Bacilli ↑, Enterobacteriales ↑, Episilonproteobacteria ↑, Gammaproteobacteria ↑, Negativicutes ↓ Order: Clostridiaceae_1 ↓, Lactobacillales ↑, Peptostreptococcaceae ↓, Selenomonadales ↓ Family: Asaccharospora ↓, Enterobacteriaceae ↑, Romboutsia ↓ Genus: Blautia ↑, Ruminiclostridium_6 ↓, Streptococcus ↑ |
NA |
| [61] | Danish | Metformin treated for 6 weeks in healthy subjects (n = 22) |
versus before metformin treatment in healthy subjects (n = 18) Genus: Bilophila ↑, Caproiciproducens ↑, Clostridium_sensu_stricto_1 ↓, Escherichia-Shigella ↑, Intestinibacter ↓, Prevotella ↑, Terrisporobacter ↓ Species: Alistipes finegoldii ↑, Bilophila wadsworthia ↑, Intestinibacter bartlettii ↓ |
NA |
3. Potential Mechanisms of Metformin on the Gut Microbiome
3.1. Regulation of Glucose Homeostasis
Based on the knowledge regarding metformin’s gut-restricted glucose-lowering effects, further investigations have been undertaken to understand the role of metformin in the gut (
)
[23]
. In particular, the upper small intestine is responsible for triggering gut peptide-dependent negative feedback signals, followed by nutrient intake
. One of the signals is glucagon-like peptide-1 (GLP-1) release via sodium-glucose cotransporter-1 (SGLT-1), which plays a dominant role in GLP-1 secretion via the transport of 3-
O
-methyl glucose
. In this regard, metformin exhibited an increase in GLP-1 secretion and SGLT1 expression in the upper small intestine, suggesting that metformin might interact with the upper small intestinal SGLT-1 mediated glucose-sensing pathway
. According to this hypothesis, the germ-free mice, physiologically used for the “microbial knockout” model, showed alterations in the glucose metabolism-related genes in the gut when the microbiota was inoculated from the healthy mice
[73]
. In addition, prebiotics and probiotics changed the gut microbiome in relation to changes in GLP-1 secretion
. Based on these relationships, Bauer et al.
[76]
demonstrated that metformin altered the upper small intestinal microbiota, resulting in the upregulation of SGLT-1 expression. Additionally, a high-fat diet in rodents reduced SGLT-1 expression, which was recovered on metformin administration
[76]
. This effect might be due to the alteration in microbiota in the upper small intestine, demonstrated by the transplantation of microbiota in the metformin-treated high-fat diet (HFD)-fed rats to untreated HFD-fed rats. In particular, the abundance of
Lactobacillus
exhibited significant recovery from dysbiosis, suggesting that
Lactobacillus
is related to SGLT-1 modification after metformin administration. This result was also observed in the metformin-treated HFD-fed mice with increased Sglt1 mRNA levels in the upper small intestine
[59]
. In addition, a previous study revealed that upregulation of SGLT-1 mediated metabolites produced by
Lactobacillus
resulted in the increased glucose uptake in Caco-2 cells, and this study supported that
Lactobacillus
might be related to the glucose modulation of metformin
[77]
. In terms of modulating the glucose-sensing pathway,
Lactobacillus
was shown to modulate the glucose-sensing machinery related to other pathways, not only for SGLT-1. When Caco-2 cells were incubated with the supernatant from the cultured
Lactobacillus
, there was an increase in the expression of the GPR120 gene, known to affect the expression of GLP-1
. Furthermore,
L. gasseri
, one of the species in the genus
Lactobacillus
, was shown to affect the long-chain acyl-CoA synthetase (ACSL)-dependent glucoregulatory fatty acid-sensing pathway
[80]
. Thus, this evidence suggests that
Lactobacillus
plays a role in modulating glucose metabolism and might be associated with the improvement of glucose parameters in rodents and humans treated with probiotic supplements containing
Lactobacillus
.
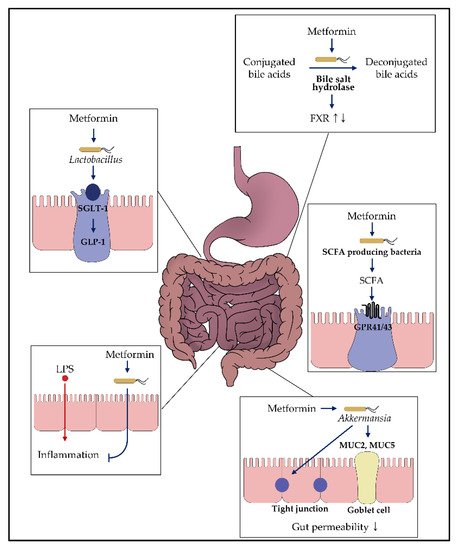
Figure 2. Impact of metformin on the gut microbiota. Various in vitro and in vivo studies demonstrated that metformin might exhibit glucose-modulating effects by interacting with the gut microbiome. Each box presents the putative mechanism suggested in this review. For more details, refer to the main text.
In conclusion, metformin recovered dysbiosis in HFD-rats, and the genus
Lactobacillus
was identified as key for modulating the glucose-sensing pathway
[76]
. However, the mechanism by which metformin alters the abundance of
Lactobacillus
remains unknown. Thus, future studies might be required to elucidate the mechanism by which metformin affects the abundance of the gut microbiota. Furthermore, alteration of
Lactobacillus
by metformin and T2DM was not consistent between the animal and human studies, as shown in
and
. For these results, Sato et al.
[28]
suggested that in human studies, the innate bacteria and bacteria originating from foods such as yogurt were not distinguished. In addition, Bauer et al.
[76]
investigated the anti-diabetic effect of metformin on the upper small intestine, comparing changes in the gut microbiome in the upper and distal intestines. Hence, these confounding factors were also regulated to unveil the relationship between metformin and the genus
Lactobacillus
.
3.2. Effects on Bacteria Producing Short-Chain Fatty Acid
Short-chain fatty acids (SCFAs), including acetate, propionate, butyrate, and lactate, are the major products of fermentation of undigestible food by the anaerobic bacteria. Based on the increasing number of studies on the relationship between the gut microbiota and metabolic disease, the effects of SCFAs produced by the gut microbiota on metabolic disease have attracted interest [81]. Indeed, SCFAs exhibit beneficial effects on glucose metabolism via multiple pathways, including activation of gut hormone receptors (e.g., Ffar2 and Ffar3) [81][82][83][84][85]. In particular, SCFAs can bind to the G protein-coupled receptor (GPR)-41 (referred to as FFAR3) and GPR-43 (referred to as FFAR2), expressed on enteroendocrine L cells, stimulating the release of GLP-1 and peptide YY that regulate glucose metabolism and insulin secretion [86][87].
Some studies have suggested that gut dysbiosis in T2DM alters the SCFA concentration. First, rodents have been used to reveal the relationship between metformin’s positive effects and SCFAs [88][89][90][91][92][93][94], specifically in the phylum Bacteroidetes, abounding in the intestine, which mainly produces acetate and propionate, imparting protective effects against insulin resistance [81][89][95][96]. The abundance of Bacteroides, one of the genera in the phylum Bacteroidetes, were observed to increase with metformin treatment in high-fat diet mice (Table 2) [88][89][90][91]. Following an increase in the abundance of Bacteroides, the concentration of SCFAs in feces of those treated with metformin was higher than that in db/db mice [91]. In vivo experiments using rodents, in vitro gut microbiome culture [97], and in silico modeling demonstrated similar results [98]. However, Brandt et al. [99] showed a negligible difference in the abundance of Bacteroides, as shown in Table 2 [89][90]. These studies used the same animal model C57BL/6J mice, but the gut microbiome was altered owing to the difference in sex, similar to the previous studies that revealed sex-dependent alterations in the gut microbiome [100][101]. Bacteroides were observed to be more abundant in female mice than in male mice. In this respect, Lee et al. [88] suggested that gut microbiota could be affected by hormone levels, subsequently influencing glucose and lipid metabolism [102][103] and one of the studies demonstrated that progesterone promotes the growth of oral Bacteroides species [104]. Although various studies have demonstrated a positive relationship between the abundance of Bacteroides and therapeutic effect of metformin, future studies should consider sexual effects to understand the effect of the hormones on Bacteroides.
Butyricimonas spp., one of the genera in the phylum Bacteroidetes, produces butyrate, a moiety known to increase insulin sensitivity [105] and regulate the gut hormones [106]. Butyricimonas spp. were increasingly abundant in metformin-treated mice [89][91]. Besides this, the abundance of genus Allobaculum, a butyrate producer [107], and Parabacteroides, producer of succinate, [108] were also increased in metformin-treated mice [89][90][109][110]. Abundant microbiota-producing SCFAs were also observed in the human fecal samples, details regarding the same are given in Section 4.
In summary, an increase in the abundance of gut microbiota-producing SCFAs might be considered as an anti-diabetic mechanism mediated by metformin treatment. Although gut microbiota producing SCFAs (e.g., the genus Allobaculum, Bacteroides, and Parabacteroides) might impart beneficial metabolic homeostasis in the host, the mechanism by which metformin affects the gut microbiota is unclear.
3.3. Enhancement of the Gut Permeability
Several studies have revealed that metabolic disorders are associated with increased gut permeability, which further increases the intestinal LPS permeability and induces chronic inflammation that causes insulin resistance [49][111][112][113]. The mucus layer plays an important role in maintaining gut permeability and gastrointestinal functions by providing substrates for bacterial growth adhesion and protection [114][115][116]. From this perspective, several studies suggest that colonization of several gut microbiota on the mucus layer induces diabetes or metabolic disorders from a dysbiosis-mediated high-fat diet [110][117][118].
Akkermansia muciniphila, belonging to the phylum Verrucomicrobia, colonizes the mucus layer of the human gastrointestinal tract and exhibits 3%–5% more microbial community in the healthy subjects than in the diabetic subjects (patients or animals) ([116][119] and references therein). A. muciniphila is an intestinal mucin-degrading bacterium that simultaneously stimulates mucin production, playing a key role in regulating glucose homeostasis in A. muciniphila [88][110][120]. Several studies have revealed that metformin treatment increases the abundance of A. muciniphila in the gut [88][89][90][91][99][109][110][121][122][123]. According to the study of Shin et al. [110], A. muiciniphila administered to HFD-fed mice showed improvement in glucose tolerance, consistent with metformin treatment in HFD-fed mice. In addition, they revealed that the proportion of A. muciniphila increased in metformin-treated HFD-mice and showed a positive correlation with the number of goblet cells producing mucin. As previously reported, an increase in the mucus layer by goblet cells might function as a barrier for LPS [49][111][112]. In this regard, Ahmadi et al. [92] suggested that metformin suppresses Wnt signaling, a critical pathway to regulate iSCs differentiation to goblet cells. In addition, these alterations were observed when the fecal microbiome was transplanted from the metformin-treated mice to control mice, suggesting that modulation of the gut microbiome by metformin is also associated with an increase in the goblet cells [92]. In this context, the expression of MUC2 and MUC5 genes, which contribute to the mucin levels, was increased in the metformin -treated HFD female mice [88]. With an increase in the expression of MUC2, several studies demonstrated that the tight-junction proteins, such as Zonulin-1 and occludin, were recovered after metformin treatment [92][94][99][124], and the intestinal permeability was reduced [94].
Table 2. Alteration of the gut microbiota-mediated metformin treatment in animal studies. ↑ (increase), ↓ (decrease), – (no alteration), NA (not applicable), Ref * (reference number).
| Ref * | Animal | Study Design | Gut Microbiota | Biochemical Alterations |
|---|---|---|---|---|
| [76] | Rats | Metformin treatment in high-fat diet | versus without metformin treatment in high fat diet Family: Lactobacillaceae ↑ Genus: Achromobacter –, Acinetobacter –, Azorhiziphilus –, Enterococcus –, Escherichia –, Klebsiella –, tobacillus ↑, Sarcina –, Stenotrophomnas – |
NA |
| [88] | Mice | Metformin treatment in high-fat diet | versus without metformin treatment in high fat diet α-diversity (Shannon): ↓ Phylum: Bacteroidetes ↑, Verrucomicrobia ↑ Family: Bacteroidaceae ↑, Clostridiales familyXIII ↑, Incertae sedis ↑, Rikenellaceae ↑, Ruminococcaceae ↑, Verrucomicrobioaceae ↑ Species: Akkermansia muciniphila ↑, Clostridium cocleatum ↑ |
Inflammation scores – |
| Metformin treatment in normal diet | versus without metformin treatment in normal diet α-diversity (Shannon): – Phylum: Bacteroidetes – Family: Rikenellaceae ↑, Ruminococcaceae ↑, Verrucomicrobioaceae ↑ Genus: Alistipes spp. ↑, Akkermansia spp. ↑, Clostridium spp. ↑ |
NA | ||
| [89] | Mice | Metformin treatment for 16 weeks in high-fat diet | versus without metformin treatment in high fat diet α-diversity (observed OTU): – Phylum: Bacteroidetes ↑, Firmicutes ↓, Verrucomicrobia ↑ Genus: Akkermansia ↑, Bacteroides ↑, Butyricimonas ↑, Parabacteroides ↑ |
IL-6 mRNA ↓ IL-1β mRNA ↓ |
| [90] | Mice | Metformin treatment for 24 weeks in high-fat diet | versus without metformin treatment in high fat diet α-diversity (Shannon): ↓ Phylum: Bacteroidetes ↑, Firmicutes ↓, Verrucomicrobia ↑ Family: Desulfovibrionaceae Genus: Akkermansia ↑, Bacteroides ↑, Christensenella ↑, Coprococcus↓, Dorea ↓, Lachnoclostridium ↓, Parabacteroides ↑, Papillibacter ↓, Oscillospira ↓, Ruminococcus ↓, Desulfovibrio ↓, Muribaculum ↓ |
Plasma threonine ↓, methionine sulfoxide ↓, Tetradecanoylcarnitine ↓, Hexadecenoylcarnitine ↓ |
| [91] | Mice | Metformin treatment in obese mice (db/db mice) | versus without metformin treatment in obese mice (db/db mice) α-diversity (Shannon): ↑ Genus: Akkermansia ↑, Butyricimonas ↑, Clostridium ↓, Coprococcus ↑, Dehalobacterium ↑, Dorea ↑, Lactobacillus ↑, Oscillospira ↑, Parabacteroides ↓, Paraprevotella ↑, Prevotella ↓, Proteus ↓, Ruminococcus ↑ |
Total SCFA concentration in feces ↑ Acetic acid ↑, Butyric acid ↑ LPS levels ↓ |
| [92] | Mice | Metformin treatment in high-fat diet | versus without metformin treatment in high fat diet α-diversity (Shannon, evenness): – Phylum: Bacteroidetes ↑ Family: Coriobacteriaceae ↓, Ruminococcaceae ↑, S24_7 ↑, Veilonellaceae ↓ Genus: Dorea ↓, Dehalobacterium ↓, Lactobacillus ↓, Lactococcus ↑, Roseburia ↓, SMB53 ↓ |
IL-6 ↓, IL-1β ↓, TNF α ↓ Taurine ↑, Butyrate ↑, Total Bile acids ↑, Propionate ↑, Leucine ↑, Creatinine ↓, Sarcosine ↓, Glutamate ↓, Pyruvate ↓, Formate ↓ |
| [93] | Rats | Metformin treatment in high-fat diet combined with a low dose streptozocin | versus without metformin treatment in high fat diet α-diversity (Simpson, Shannon): ↑ Class: Coriobacteriia ↑ Family: S24_7 ↑ |
Total SCFAs ↑, Butyric acid ↑, Isovaleric acid ↑ |
| [94] | Rats | Metformin treatment in high-fat diet combined with a low dose streptozocin | versus without metformin treatment in high fat diet α-diversity (Chao1): ↑ Family: S24_7 ↓ Genus: Anaerotruncus ↑, Escherichia-Shiegella ↓, Eubacterium xylanophilum ↑, Lachnospiraceae NK4A136 ↑, Lachnospiraceae-UCG_006 ↑, Roseburia ↑ |
Serum LPS ↓, Serum CRP↓, Serum TNF α ↓, Serum IL-6 ↓ Propionate in cecum ↑, Butyrate in cecum ↑ |
| [99] | Mice (female) |
Metformin treatment in fat, fructose and cholesterol rich diet |
versus without metformin treatment in fructose and cholesterol rich diet Family: Alloprevotella ↓ Genus: Bacteroides –, Romboutsia ↓ Species: Akkermansia muciniphila –, Lactobacillus animalis ↓ |
TNF α ↓ Endotoxin ↓ |
| [109] | Mice | Metformin treatment for 5 weeks in high-fat diet combined with a low dose streptozocin |
versus without metformin treatment in high fat diet α-diversity (observed OTU): – Genus: Akkermansia ↑, Bacteroides spp. ↓ |
NA |
| [110] | Mice | Metformin treatment in high-fat diet | versus without metformin treatment in high fat diet Phylum: Verrucomicrobia ↑, Genus: Akkermansia ↑, Alistipes ↑, Anaerotruncus ↓, Blautia ↓, Lactococcus ↓, Lactonifactor ↓, Lawsonia ↓, Odoribacter ↓, Parabacteroides ↓ |
IL-6 mRNA ↓ IL-1β mRNA ↓ |
| [121] | Mice | Metformin treatment for 30 days |
versus without metformin treatment in healthy mice α-diversity (Shannon): – Class: Lachnopiraceae ↓, Porphyromonadaceae ↑, Prevoltellaceae ↑, Rhodobacteraceae ↓, Rikenellaceae ↑, Verrucomicrobiaceae ↑ |
NA |
| [123] | Rats | Metformin treatment in high-fat diet |
versus without metformin treatment in high fat diet α-diversity (Shannon): ↓ Phylum: Bacteroidetes –, Firmicutes –, Proteobacteria ↑ Species: Akkermansia ↑, Allobaculum ↑, Bacteroides ↑, Blautia ↑, Butyricoccus ↑, Clostridium ↓, Klebsiella ↑, Lactobacillus ↑, Parasutterella ↑, Phascolarctobacterium ↑, Prevotella ↑, Roseburia ↓ |
NA |
| [124] | Rats | Metformin treatment in high-fat diet combined with a low dose streptozocin |
versus without metformin treatment in high fat diet α-diversity (Chao1, Shannon): ↑ Phylum: Bacteroidetes ↑, Firmicutes ↓, Proteobacteria ↓ Order: Clostridiales ↑, Enterobacteriales ↓, Lactobacillales ↑ Genus: Akkermansia ↑, Desulfovibrio ↓, Lachnospiraceae NK4A136 ↓, Lactobacillus ↑, Roseburia ↑ |
NA |
| [125] | Mice | Metformin treatment for 3 weeks in high-fat diet | versus without metformin treatment in high fat diet α-diversity (Shannon, evenness): – Genus: Akkermansia ↑, Allobaculum ↓, Clostridium ↓, Enterococcus ↓, Lactococcus ↓, Leuconostoc ↓, Oscillospira ↑, Parabacteroides ↑, Prevotella ↑, Ruminococcus ↓, Streptococcus ↓ |
NA |
| [126] | Mice with | Metformin treatment for 5 weeks in high-fat diet combined with a low dose streptozocin |
versus without metformin treatment in high fat diet α-diversity (Chao1): ↓ Phylum: Bacteroidetes ↓, Firmicutes ↑, Proteobacteria ↓ Genus: Lactobacillus ↑ |
NA |
| [127] | Mice | Metformin treatment in high-fat diet |
versus without metformin treatment in high fat diet α-diversity (Shannon, evenness): – Species: Bacteriodetes fragilis ↓, Escherichia coli ↓ |
Serum endotoxin ↓ IL-6 ↓, TLR4 ↓ |
| [128] | Rats | Metformin treatment in high-fat diet combined with a low dose streptozocin |
versus without metformin treatment in high fat diet α-diversity (Chao1): ↑ Phylum: Bacteroidetes ↑, Proteobacteria ↓, Verrucomicrobia ↓ Family: Alcaligenaceae ↑, Peptococcaceae ↑, Prevotellaceae ↑, S24_7 ↑ Genus: Prevotella ↑, Sutterella ↑, 02d06 ↑, rc4 ↑ |
IL-6 mRNA in pancrease ↓, TNF α mRNA in pancrease ↓, LPS ↓ |
| [129] | Rats | Metformin treatment in high-fat diet combined with a low dose streptozocin |
versus without metformin treatment in high fat diet Genus: Bifidobacterium ↑, Lactobacillus ↑ Species: Clostridium perfringens ↓, Escherichia coli ↓ |
Plasma endotoxin ↓, Total SCFAs in cecum ↑, Lactic acid in cecum ↑, Acetic acid in cecum ↑ |
| [130] | Rats | Metformin treatment in Otsuka Long-Evans Tokushima Fatty (OLETF) rats |
versus without metformin treatment Genus: Akkermansia ↑, Prevotella ↓, Roseburia ↑ Species: Escherichia coli ↓ |
Serum endotoxin ↓, Fecal endotoxin ↓, serum TNF α ↓, serum IL-6 ↓ |
| [131] | Rats | Metformin treatment in Zucker diabetic fatty rats | versus without metformin treatment α-diversity (Shannon): – Phylum: Bacteroidetes –, Firmicutes –↑, Proteobacteria ↓, Tenericutes –, Verrucomicrobia ↑ Genus: Lactobacillus ↑ Species: Lactobacillus intestinalis ↑, Lactobacillus johnsonii ↑ |
NA |
Furthermore, A. muciniphila has been shown to strengthen the intestinal barrier by increasing the expression of the tight junction proteins [132][133][134]. Based on this supporting evidence, metformin might be capable of reducing gut permeability via increased expression of the mucin and tight-junction proteins. However, Shin et al. [110] revealed that there was no substantial change in the gut permeability of LPS and suggested that increased goblet cells may act as a barrier against LPS by producing immune-effector molecules [135][136]. These differences among studies might be due to the duration of treatment, dosage, or the amount of A. muciniphila in the gut. In addition, the in vitro culture [132][133][134] using bacteria is complex; thus, it might be different from the in vivo studies [88][89][90][91][99][109][110][121][122][123].
In conclusion, the increase in the abundance of A. muciniphila by metformin treatment promotes mucin production, which might recover the increased gut permeability induced by high-fat diets or metabolic disorders. However, until now, the direct effects of metformin on cellular pathways to increase A. muciniphila remain unclear. Thus, further investigation to identify the physiological pathway by which metformin increases A. muciniphila will help understand the unidentified effects of metformin on the gut microbiota.
3.4. Modulation of the Immune Response
In the past decade, accumulating evidence from a variety of animal models or clinical studies has shown that metabolic disorders, including T2DM, are associated with chronic or subacute tissue inflammation in the adipose tissue and liver, causing insulin resistance [137][138][139][140][141]. Several studies in T2DM, have reported that metformin modulates inflammation via inflammatory modulating signaling pathways, such as STAT3 signaling [142] or the NF-κB (nuclear factor kappa light chain enhancer of activated B cells) signaling pathway [94][143][144]. Metformin directly suppresses the release of an inflammatory cytokine such as interleukin 6 (IL-6), interleukin 1β (IL-1β), and TNF-α (tumor necrosis factor– α) [92][127][128]. Thus, several studies have reported alterations in the expression of IL-6 following metformin treatment, and some reports have even showed that the alteration of microbiota due to metformin treatment was related to the modulation of inflammation.
In detail, A. muciniphila, the abundance of which increased upon metformin treatment as mentioned above, also exhibited anti-inflammatory effects in the gut, consistent with previous studies that revealed the anti-inflammatory effect of A. muciniphila [110][145][146]. Shin et al. [110] demonstrated that decreased regulatory T cells, a regulator of immune responses, in the stromal vascular fraction of the HFD-control mice was recovered by A. muciniphila and metformin treatment. Furthermore, the IL-6 and IL-1β mRNA levels were significantly decreased in A. muciniphila on metformin treatment [110]. In this context, a negative correlation can be drawn between the abundance of A. muciniphila upon metformin treatment and inflammatory markers, such as inflammatory cytokines or LPS concentration [89][91][121][122]. These effects of A. muciniphila on inflammation have also been demonstrated in human studies (Clinical Trials.gov Identifier: NCT02637115) with fewer inflammation markers and improved insulin sensitivity [147].
Likewise, the abundance of Bacteroides and Butyricimonas also increased upon metformin treatment [89][90][91][97][98][109][123]. In particular, Lee et al. [89] revealed that IL-6 expression negatively correlated with the abundance of Bacteroides and Butyricimonas. Above all, IL-6 possesses not only pro-inflammatory effects but also attenuates insulin signaling in adipocytes [148][149][150]. Thus, decreased IL-6 expression on metformin treatment may contribute to its anti-diabetic effect. In addition, Lee et al. [89] showed that the expression of IL-1β, which is related to insulin resistance, decreased while the abundance of Bacteroides and Butyricimonas was increased. The tendency to decrease the expression of IL-6, IL-1β and TNF-α was also observed in other studies, but the types of bacteria that correlated with the expression of inflammatory cytokines were different [92][94][128][130]. With the inhibition of pro-inflammatory cytokines, modulation of the inflammatory signaling pathway is a potential mechanism to attenuate inflammation. The TLR/NF-κB signaling pathway also plays a role in intestinal inflammation [151]. Zhang et al. [91] demonstrated that the metformin-treated group exhibited downregulation of the intestinal TLR/NF-κB signaling activities. A similar result was observed wherein phosphorylation of IKKα/β upstream of NF-κB signaling was decreased in metformin-treated mice [99]. In addition, abundance of other gut bacteria increased in the metformin-treated group and were known to interact with the host immune response. For example, Roseburia is more abundant in the metformin-treated group and is known to inhibit the activity of NF-κB [92][94][124][130][152][153]. In addition, the genus Lactobacillus and several Lactobacillus species have been shown to modulate inflammation, as reported in previous studies [154][155][156][157]. Thus, future studies should be warranted to unveil how metformin prevents the host inflammatory response related to the alteration of gut bacteria.
To conclude, various inflammatory markers were correlated with the alteration of bacteria on metformin treatment. Furthermore, these effects have also been supported by other studies that demonstrated the therapeutic effects of metformin on inflammatory diseases (e.g., non-alcoholic fatty liver disease and polycystic ovary syndrome) through interaction with the gut microbiota [99][158].
3.5. Actions on the Circulation of the Bile Acids
Bile acids are synthesized from cholesterol in the liver and secreted into the intestine, following which cholic acid and chenodeoxycholic acid are converted to secondary bile acids, such as deoxycholic acid and lithocholic acid, via enzymes and gut microbiota. For several decades, bile acids have been shown to play a role in glucose, lipid, and energy metabolism [159]. The modulation effects of bile acids on several metabolic pathways are mainly via binding to several intracellular nuclear receptors, including farnesoid X receptor (FXR), pregnane X receptor (PXR), and cell surface G protein-coupled receptors (GPCRs) ([160] and references therein). In this regard, metformin showed an inhibitory effect on the bile acid resorption, resulting in increased exposure of the gut to bile acids [92][161][162][163]. Napolitano et al. [55] demonstrated the effect of metformin on bile acid in a clinical trial in T2DM patients. Extended exposure to bile acid might allow bile acids to bind to the intestinal FXR. Thus, the glucose-modulating effect of metformin via bile acids seems to be related to the FXR signaling. However, the glucose-modulating effect mediated by FXR remains controversial. There is some evidence that inactivation of FXR results in better glucose control and increased GLP-1 secretion [164][165][166]. For example, FXR-deficient mice exhibit increased GLP-1 expression and improved glucose metabolism [164]. In contrast to these results, some studies suggested that activation of FXR via FXR agonists improves glucose tolerance and insulin sensitivity [167][168][169][170][171]. Thus, the glucose-modulating effect of metformin via bile acid circulation has not yet been clarified. Recently, a study revealed that metformin acts on the B. fragilis-glycoursodeoxycholic acid (GUDCA)-intestinal FXR axis, improving hyperglycemia [59]. GUDCA, a conjugated bile acid, is deconjugated by the gut microbiome and is demonstrated to be an FXR antagonist. Sun et al. [59] revealed that metformin inhibited the deconjugation of GUDCA through the activity of the bile salt hydrolase of B. fragilis, resulting in an increased GUDCA concentration. This result is consistent with the correlation between GUDCA levels in stool and the presence of B. fragilis. Additionally, the abundance of Lactobacillus sanfrancisensis, contained in the genus Lactobacillus known to affect intestinal FXR signaling, was increased in the metformin-treated HFD-fed mice [80].
Taken together, metformin has a role in modulating glucose homeostasis via the regulation of the bile acid circulation. Conflict in the function of FXR in glucose homeostasis might be due to different agonists and antagonists for FXR (e.g., intestinal FXR agonist or whole-body FXR agonist) [160]. Furthermore, bile acid pools in mice and humans are known to be quite different and might have a conflicting role in FXR. As a result, further studies could be conducted by considering these confounding factors.
4. Relationships between Metformin and Gut Microbiome in Human Studies
The glucose-modulating effect of metformin on the gut microbiome has been evaluated in various clinical trials. The first clinical study that observed the relationship between metformin and the gut microbiome was conducted as an open-label, single-group study in T2DM patients
[55]
. In this study, they demonstrated alterations in the composition of the gut microbiome, glucose hormone, glucose-related parameters, and bile acid concentration in feces. A similar tendency was observed in the present study, despite minor differences in the gut microbiome composition (
).
First, at the phylum level, the alterations in the abundance of Firmicutes and Bacteroidetes were remarkable on comparing visits 3 (non-treatment) and 4 (metformin treatment). Although there were differences among subjects, the abundance of Firmicutes was commonly increased, whereas that of Bacteroidetes was decreased after metformin treatment. This result is in line with the previous finding that the Firmicutes/Bacteroidetes ratios were considered a predictor for metabolic disease such as T2DM or obesity in several human studies
. The Firmicutes/Bacteroidetes ratios were decreased in the T2DM patients, and this phenomenon was recovered by metformin treatment in several clinical studies
. In contrast to these results, some studies did not show an alteration in the ratio of Firmicutes to Bacteroidetes
[60]
. Inconsistencies among studies could be considered for the following reasons. The phyla Firmicutes and Bacteroidetes are the most abundant bacteria in the human gut and include a large number of bacterial species. Thus, a comparison of the Firmicutes and Bacteroidetes ratio is considered too simple to evaluate metabolic disease or improvement. In addition, the difference might be attributed to the compositional difference between the stool and biopsy specimens. Clinical studies in this review used fecal samples to analyze the gut microbiome, but previous studies have shown differences in the microbial compositions of biopsy and fecal samples
. In particular, the mucosa-associated microbiota, to which the phylum Firmicutes is enriched, exhibited compositional differences in biopsies derived from colon and stool samples
. Thus, Hollister et al.
[174]
suggested biopsy or surgical specimens for the evaluation of mucosa-associated microbiota. For these reasons, further investigations require the validity of the Firmicutes/Bacteroidetes ratio as a relevant marker for metabolic diseases. At the genus level, it is noteworthy that
Escherichia,
including
Escherichia/Shigella
and
Escherichia coli
, exhibited a significant increase in T2DM patients upon metformin treatment. An increase in the abundance of
Escherichia/Shigella
upon metformin treatment was also observed in the other clinical trials including healthy volunteers
. Forslund et al.
[31]
suggested that metformin administration creates a competitive environment for
Escherichia
coli using nitrate or other energy sources, resulting in changes in the abundance of the gut microbiome
[177]
. Wu et al.
[56]
also demonstrated a change in the abundance of
E. coli
as an indirect effect of metformin treatment in the in vitro gut simulation. Elbere et al.
[60]
demonstrated that the abundance of
Escherichia
/
Shigella
before metformin treatment is associated with side effects. In this study, the increased presence of
Escherichia/Shigella
showed mild and severe side effects; however, this level was lower than the detection limit in the no-side-effect group. Thus, increased abundance of
Escherichia
was considered as a marker for the gastrointestinal side effects of metformin. Forslund et al.
[31]
suggested that side effects derived from
Escherichia
are due to an increase in lipopolysaccharide synthesis or sulfate metabolism potential, known to contribute to intestinal bloating
.
In contrast, the abundance of
Intestinibacter
spp. decreased in T2DM patients treated with metformin in several clinical studies
. Until now, the role of
Intestinibacter
is still unclear, Forslund et al.
[31]
suggested that
Intestinibacter
showed resistance to oxidative stress and degradation of fucose, indicating indirect mucus degradation through analysis of SEED (
, accessed on 30 March 2021)
[182]
and gut microbial modules (GMM) functional annotations.
In addition,
A. muciniphila
, which is positively correlated with metformin treatment, showed a less clear link in human studies. Although Wu et al.
[56]
demonstrated an increase in
A. muciniphila
in the in vitro pure cultures, there was no correlation between the abundance of
A. muciniphila
and % hemoglobin A1c. Furthermore, clinical studies in healthy volunteers showed no change in the abundance of
A. muciniphila
when they were treated with metformin
. The reasons for these differences might be considered to be affected by factors dependent on individuals, such as fibers
[183]
, polyphenol availability
, immune response
, and age
. Thus, it might be difficult to conclude the role of
A. muciniphila
in humans as a major contributor to the anti-diabetic effect of metformin, although improvements in the metabolic parameters were observed in the
A. muciniphila
-treated human studies.
From the perspective of biochemical alterations upon metformin treatment, there were some differences in the bile acid and SCFA concentrations in the feces. They found that metformin exposure increased the excretion of bile acid in feces, consistent with the inhibitory effect of metformin on the resorption of bile acids
. In addition, the abundance of Firmicutes and Bacteroidetes correlated with the bile acid concentration and gut peptide, suggesting that metformin indirectly regulates the secretion of gut hormones via bile acid metabolism. Increased SCFA concentration in feces or an increase in the abundance of SCFA-producing bacteria has been observed in human studies
. In particular, Wu et al.
[56]
only demonstrated that the concentration of SCFA in fecal samples, resulting in formation of butyrate and propionate, substantially increased on metformin treatment. This result is consistent with animal studies that showed that metformin increases SCFA-producing bacteria
. Thus, these clinical results support the hypothesis that metformin exerts beneficial effects via bile acids and SCFAs.
The clinical studies discussed in this review exhibited differences among studies, including observed taxonomic groups in metformin treatment and diversity in abundance. As far as diversity is concerned, only a few subjects were engaged in the clinical study, resulting in no statistical difference in the diversity of the gut microbiome. This issue has been inconsistent in clinical trials (
). Forslund et al.
[31]
conducted a meta-analysis of metagenomic data from Swedish, Danish, and Chinese individuals. In this study, gut microbiome was less rich T2DM patients without metformin treatment; this richness slightly recovered, almost as much as that in the control group, in T2DM patients on metformin treatment
[31]
. The diversity was also shown to decrease in the Chinese T2DM patients
[79]
, which was consistent with the results from the study by Forslund et al.
[31]
.
These differences might be derived from the dosage, study duration, disease state, race differences, and sample size. Thus, to elucidate the anti-diabetic effect of metformin via modulation of the gut microbiome, clinical studies in ethnic-controlled environments or comparisons among ethnicities are required. Indeed, clinical studies in various populations have been conducted at
(assessed on 30 March 2021) (
). Most of the research to date has revealed taxonomic groups in the gut at the genus level, and not at the species level, due to technical limitations. To counter this limitation, recent studies introduced a gut microbiome analysis method to make a possible profile at the species level
. In the future, these methods to analyze the gut microbiome could help clarify the relationship between metformin and the gut microbiome.
Table 3.
Enrolled clinical studies to investigate the relationship between the gut microbiome and metformin in recruiting or active state.
| Clinical Trials.gov Identifier | Study Title | Country | Study Population | Interventions |
|---|---|---|---|---|
| NCT04194515 | Gut Microbiota and Bile Acids in Type 2 Diabetes Mellitus | Taiwan | Outpatients and treatment-naïve male patients with type 2 diabetes |
Drug: YH1 Drug: metformin |
| NCT04287387 | Response of Gut Microbiota in Type 2 Diabetes to Hypoglycemic Agents | China | Type 2 diabetes patients (18–65 years) |
Drug: Glucophage 500 mg Tablet Drug: Acarbose Tablets Drug: Sitagliptin tablet Drug: Dapagliflozin Tablet Drug: Pioglitazone Tablets Drug: Glimepiride Tablets |
| NCT04639492 | Postbiotic MBS and Metformin Combination in Patients With T2DM |
Taiwan | Type 2 diabetes patients (20–70 years) |
Dietary Supplement: MBS oral solution Oral BIDAC, twice a day before breakfast and dinner times |
| NCT02960659 | Title: Therapeutic Targets in African-American Youth With Type 2 Diabetes |
United States | African-American (12–25 years) |
Drug: Metformin and Liraglutide Drug: Metformin |
| NCT03558867 | Personalized Medicine in Pre-diabetes and Early Type 2 Diabetes |
Australia | Pre-diabetes or newly-diagnosed with type 2 diabetes (in the last 6 months) |
Drug: Metformin + Healthy diet Drug: Metformin + Personalized diet |
| NCT03732690 | The Interaction Between Protein Intake, Gut Microbiota and Type 2 Diabetes in Subjects With Different Ethnic Backgrounds |
France | T2DM patients: Caucasian (n = 80), Caribbean (n = 40) stable dose of metformin and do not use insulin or proton-pump inhibitors. |
Other: Diet HP Other: Diet LP |
| NCT04089280 | Probiotics in Metformin Intolerant Patients With Type 2 Diabetes | Poland | T2DM patients (18–75 years) with metformin treatment in the last 3 months (<1500 mg/d) | Dietary Supplement: Sanprobi Barrier-multispecies probiotic Other: Placebo Comparator |
| NCT03718715 | The Interaction Between Metformin and Microbiota—The MEMO Study. (MEMO) |
Sweden | Newly diagnosed patients with type 2 diabetes without previous treatment with metformin (40–80 years). |
Drug: Metformin |
| NCT03489317 | Gut Microbiomes in Patients With Metabolic Syndrome |
Hongkong | Residents in Hongkong (no metabolic syndrome, metabolic syndrome-partial, metabolic syndrome-full) | Drug: Metformin Behavioral: lifestyle modification Drug: Simvastatin 10 mg Drug: Amlodipine 5 mg |
| NCT02609815 | Initial Combination of Gemigliptin and Metformin on Microbiota Change |
Republic of Korea | Type 2 patients with drug naive for 6 weeks | Drug: gemigliptin/metformin Drug: glimepiride/metformin |
| NCT04341571 | Effect of Probiotics Versus Metformin on Glycemic Control, Insulin Sensitivity and Insulin Secretion in Prediabetes. |
Mexico | Pre-diabetes | Dietary Supplement: Probiotics Drug: Metformin |
| NCT04209075 | Prebiotics and Metformin Improve Gut and Hormones in Type 2 Diabetes in Youth (MIGHTY-fiber) | United States | Type 2 patients (10–25 years) | Dietary Supplement: Biomebliss Drug: Metformin Dietary Supplement: Placebo |
References
- Foretz, M.; Guigas, B.; Bertrand, L.; Pollak, M.; Viollet, B. Metformin: From mechanisms of action to therapies. Cell Metab. 2014, 20, 953–966.
- Andujar-Plata, P.; Pi-Sunyer, X.; Laferrere, B. Metformin effects revisited. Diabetes Res. Clin. Pract. 2012, 95, 1–9.
- Rena, G.; Pearson, E.R.; Sakamoto, K. Molecular mechanism of action of metformin: Old or new insights? Diabetologia 2013, 56, 1898–1906.
- Rojas, L.B.; Gomes, M.B. Metformin: An old but still the best treatment for type 2 diabetes. Diabetol. Metab. Syndr. 2013, 5, 6.
- Viollet, B.; Guigas, B.; Sanz Garcia, N.; Leclerc, J.; Foretz, M.; Andreelli, F. Cellular and molecular mechanisms of metformin: An overview. Clin. Sci. 2012, 122, 253–270.
- Graham, G.G.; Punt, J.; Arora, M.; Day, R.O.; Doogue, M.P.; Duong, J.K.; Furlong, T.J.; Greenfield, J.R.; Greenup, L.C.; Kirkpatrick, C.M.; et al. Clinical pharmacokinetics of metformin. Clin. Pharmacokinet. 2011, 50, 81–98.
- Beckmann, R. [Absorption, distribution in the organism and elimination of metformin]. Diabetologia 1969, 5, 318–324.
- Wilcock, C.; Wyre, N.D.; Bailey, C.J. Subcellular distribution of metformin in rat liver. J. Pharm. Pharmacol. 1991, 43, 442–444.
- Rena, G.; Hardie, D.G.; Pearson, E.R. The mechanisms of action of metformin. Diabetologia 2017, 60, 1577–1585.
- Wang, D.S.; Jonker, J.W.; Kato, Y.; Kusuhara, H.; Schinkel, A.H.; Sugiyama, Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 2002, 302, 510–515.
- Shu, Y.; Sheardown, S.A.; Brown, C.; Owen, R.P.; Zhang, S.; Castro, R.A.; Ianculescu, A.G.; Yue, L.; Lo, J.C.; Burchard, E.G.; et al. Effect of genetic variation in the organic cation transporter 1 (oct1) on metformin action. J. Clin. Investig. 2007, 117, 1422–1431.
- Natali, A.; Ferrannini, E. Effects of metformin and thiazolidinediones on suppression of hepatic glucose production and stimulation of glucose uptake in type 2 diabetes: A systematic review. Diabetologia 2006, 49, 434–441.
- Foretz, M.; Hebrard, S.; Leclerc, J.; Zarrinpashneh, E.; Soty, M.; Mithieux, G.; Sakamoto, K.; Andreelli, F.; Viollet, B. Metformin inhibits hepatic gluconeogenesis in mice independently of the lkb1/ampk pathway via a decrease in hepatic energy state. J. Clin. Investig. 2010, 120, 2355–2369.
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014, 510, 542–546.
- Bonora, E.; Cigolini, M.; Bosello, O.; Zancanaro, C.; Capretti, L.; Zavaroni, I.; Coscelli, C.; Butturini, U. Lack of effect of intravenous metformin on plasma concentrations of glucose, insulin, c-peptide, glucagon and growth hormone in non-diabetic subjects. Curr. Med. Res. Opin. 1984, 9, 47–51.
- Stepensky, D.; Friedman, M.; Raz, I.; Hoffman, A. Pharmacokinetic-pharmacodynamic analysis of the glucose-lowering effect of metformin in diabetic rats reveals first-pass pharmacodynamic effect. Drug Metab. Dispos. 2002, 30, 861–868.
- Bailey, C.J.; Mynett, K.J.; Page, T. Importance of the intestine as a site of metformin-stimulated glucose utilization. Br. J. Pharmacol. 1994, 112, 671–675.
- Bailey, C.J.; Wilcock, C.; Scarpello, J.H. Metformin and the intestine. Diabetologia 2008, 51, 1552–1553.
- Tucker, G.T.; Casey, C.; Phillips, P.J.; Connor, H.; Ward, J.D.; Woods, H.F. Metformin kinetics in healthy subjects and in patients with diabetes mellitus. Br. J. Clin. Pharmacol. 1981, 12, 235–246.
- Wilcock, C.; Bailey, C.J. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica 1994, 24, 49–57.
- Jensen, J.B.; Sundelin, E.I.; Jakobsen, S.; Gormsen, L.C.; Munk, O.L.; Frokiaer, J.; Jessen, N. [11c]-labeled metformin distribution in the liver and small intestine using dynamic positron emission tomography in mice demonstrates tissue-specific transporter dependency. Diabetes 2016, 65, 1724–1730.
- Dujic, T.; Zhou, K.; Donnelly, L.A.; Tavendale, R.; Palmer, C.N.; Pearson, E.R. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: A godarts study. Diabetes 2015, 64, 1786–1793.
- Buse, J.B.; DeFronzo, R.A.; Rosenstock, J.; Kim, T.; Burns, C.; Skare, S.; Baron, A.; Fineman, M. The primary glucose-lowering effect of metformin resides in the gut, not the circulation: Results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care 2016, 39, 198–205.
- Kho, Z.Y.; Lal, S.K. The human gut microbiome—A potential controller of wellness and disease. Front. Microbiol. 2018, 9, 1835.
- Hooper, L.V.; Gordon, J.I. Commensal host-bacterial relationships in the gut. Science 2001, 292, 1115–1118.
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546.
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60.
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut dysbiosis and detection of “live gut bacteria” in blood of japanese patients with type 2 diabetes. Diabetes Care 2014, 37, 2343–2350.
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in european women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103.
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085.
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266.
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108.
- Zhang, F.; Wang, M.; Yang, J.; Xu, Q.; Liang, C.; Chen, B.; Zhang, J.; Yang, Y.; Wang, H.; Shang, Y.; et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 2019, 66, 485–493.
- Ahmad, A.; Yang, W.; Chen, G.; Shafiq, M.; Javed, S.; Ali Zaidi, S.S.; Shahid, R.; Liu, C.; Bokhari, H. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLoS ONE 2019, 14, e0226372.
- Chavez-Carbajal, A.; Pizano-Zarate, M.L.; Hernandez-Quiroz, F.; Ortiz-Luna, G.F.; Morales-Hernandez, R.M.; De Sales-Millan, A.; Hernandez-Trejo, M.; Garcia-Vite, A.; Beltran-Lagunes, L.; Hoyo-Vadillo, C.; et al. Characterization of the gut microbiota of individuals at different t2d stages reveals a complex relationship with the host. Microorganisms 2020, 8, 94.
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590.
- Rodriguez, J.; Hiel, S.; Delzenne, N.M. Metformin: Old friend, new ways of action-implication of the gut microbiome? Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 294–301.
- Wu, T.; Horowitz, M.; Rayner, C.K. New insights into the anti-diabetic actions of metformin: From the liver to the gut. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 157–166.
- Hur, K.Y.; Lee, M.S. New mechanisms of metformin action: Focusing on mitochondria and the gut. J. Diabetes Investig. 2015, 6, 600–609.
- Vallianou, N.G.; Stratigou, T.; Tsagarakis, S. Metformin and gut microbiota: Their interactions and their impact on diabetes. Hormones 2019, 18, 141–144.
- Zhang, Q.; Hu, N. Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 2020, 13, 5003–5014.
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79.
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031.
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484.
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and firmicutes/bacteroidetes ratio in an adult ukrainian population. BMC Microbiol. 2017, 17, 120.
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojarvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913–916.e7.
- Halawa, M.R.; El-Salam, M.A.; Mostafa, B.M.; Sallout, S.S. The gut microbiome, lactobacillus acidophilus; relation with type 2 diabetes mellitus. Curr. Diabetes Rev. 2019, 15, 480–485.
- Zeuthen, L.H.; Christensen, H.R.; Frokiaer, H. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin. Vaccine Immunol. 2006, 13, 365–375.
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772.
- Delzenne, N.M.; Cani, P.D.; Everard, A.; Neyrinck, A.M.; Bindels, L.B. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia 2015, 58, 2206–2217.
- Carvalho, B.M.; Guadagnini, D.; Tsukumo, D.M.L.; Schenka, A.A.; Latuf-Filho, P.; Vassallo, J.; Dias, J.C.; Kubota, L.T.; Carvalheira, J.B.C.; Saad, M.J.A. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 2012, 55, 2823–2834.
- Song, M.J.; Kim, K.H.; Yoon, J.M.; Kim, J.B. Activation of toll-like receptor 4 is associated with insulin resistance in adipocytes. Biochem. Biophys. Res. Commun. 2006, 346, 739–745.
- Balakumar, M.; Prabhu, D.; Sathishkumar, C.; Prabu, P.; Rokana, N.; Kumar, R.; Raghavan, S.; Soundarajan, A.; Grover, S.; Batish, V.K.; et al. Improvement in glucose tolerance and insulin sensitivity by probiotic strains of indian gut origin in high-fat diet-fed c57bl/6j mice. Eur. J. Nutr. 2018, 57, 279–295.
- Panwar, H.; Rashmi, H.M.; Batish, V.K.; Grover, S. Probiotics as potential biotherapeutics in the management of type 2 diabetes—Prospects and perspectives. Diabetes Metab. Res. Rev. 2013, 29, 103–112.
- Napolitano, A.; Miller, S.; Nicholls, A.W.; Baker, D.; Van Horn, S.; Thomas, E.; Rajpal, D.; Spivak, A.; Brown, J.R.; Nunez, D.J. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS ONE 2014, 9, e100778.
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858.
- De la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velasquez-Mejia, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 2017, 40, 54–62.
- Huang, F.; Nilholm, C.; Roth, B.; Linninge, C.; Hoglund, P.; Nyman, M.; Ohlsson, B. Anthropometric and metabolic improvements in human type 2 diabetes after introduction of an okinawan-based nordic diet are not associated with changes in microbial diversity or scfa concentrations. Int. J. Food Sci. Nutr. 2018, 69, 729–740.
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal fxr mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929.
- Elbere, I.; Kalnina, I.; Silamikelis, I.; Konrade, I.; Zaharenko, L.; Sekace, K.; Radovica-Spalvina, I.; Fridmanis, D.; Gudra, D.; Pirags, V.; et al. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS ONE 2018, 13, e0204317.
- Bryrup, T.; Thomsen, C.W.; Kern, T.; Allin, K.H.; Brandslund, I.; Jorgensen, N.R.; Vestergaard, H.; Hansen, T.; Hansen, T.H.; Pedersen, O.; et al. Metformin-induced changes of the gut microbiota in healthy young men: Results of a non-blinded, one-armed intervention study. Diabetologia 2019, 62, 1024–1035.
- Lam, T.K. Neuronal regulation of homeostasis by nutrient sensing. Nat. Med. 2010, 16, 392–395.
- Duca, F.A.; Bauer, P.V.; Hamr, S.C.; Lam, T.K. Glucoregulatory relevance of small intestinal nutrient sensing in physiology, bariatric surgery, and pharmacology. Cell Metab. 2015, 22, 367–380.
- Gorboulev, V.; Schurmann, A.; Vallon, V.; Kipp, H.; Jaschke, A.; Klessen, D.; Friedrich, A.; Scherneck, S.; Rieg, T.; Cunard, R.; et al. Na(+)-d-glucose cotransporter sglt1 is pivotal for intestinal glucose absorption and glucose-dependent incretin secretion. Diabetes 2012, 61, 187–196.
- Gribble, F.M.; Williams, L.; Simpson, A.K.; Reimann, F. A novel glucose-sensing mechanism contributing to glucagon-like peptide-1 secretion from the glutag cell line. Diabetes 2003, 52, 1147–1154.
- Kuhre, R.E.; Frost, C.R.; Svendsen, B.; Holst, J.J. Molecular mechanisms of glucose-stimulated glp-1 secretion from perfused rat small intestine. Diabetes 2015, 64, 370–382.
- Moriya, R.; Shirakura, T.; Ito, J.; Mashiko, S.; Seo, T. Activation of sodium-glucose cotransporter 1 ameliorates hyperglycemia by mediating incretin secretion in mice. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1358–E1365.
- Parker, H.E.; Adriaenssens, A.; Rogers, G.; Richards, P.; Koepsell, H.; Reimann, F.; Gribble, F.M. Predominant role of active versus facilitative glucose transport for glucagon-like peptide-1 secretion. Diabetologia 2012, 55, 2445–2455.
- Maida, A.; Lamont, B.J.; Cao, X.; Drucker, D.J. Metformin regulates the incretin receptor axis via a pathway dependent on peroxisome proliferator-activated receptor-alpha in mice. Diabetologia 2011, 54, 339–349.
- Vardarli, I.; Arndt, E.; Deacon, C.F.; Holst, J.J.; Nauck, M.A. Effects of sitagliptin and metformin treatment on incretin hormone and insulin secretory responses to oral and “isoglycemic” intravenous glucose. Diabetes 2014, 63, 663–674.
- Duca, F.A.; Cote, C.D.; Rasmussen, B.A.; Zadeh-Tahmasebi, M.; Rutter, G.A.; Filippi, B.M.; Lam, T.K. Metformin activates a duodenal ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 2015, 21, 506–511.
- Lenzen, S.; Lortz, S.; Tiedge, M. Effect of metformin on sglt1, glut2, and glut5 hexose transporter gene expression in small intestine from rats. Biochem. Pharmacol. 1996, 51, 893–896.
- El Aidy, S.; Merrifield, C.A.; Derrien, M.; van Baarlen, P.; Hooiveld, G.; Levenez, F.; Dore, J.; Dekker, J.; Holmes, E.; Claus, S.P.; et al. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 2013, 62, 1306–1314.
- Ejtahed, H.S.; Mohtadi-Nia, J.; Homayouni-Rad, A.; Niafar, M.; Asghari-Jafarabadi, M.; Mofid, V. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition 2012, 28, 539–543.
- Yadav, H.; Jain, S.; Sinha, P.R. Antidiabetic effect of probiotic dahi containing lactobacillus acidophilus and lactobacillus casei in high fructose fed rats. Nutrition 2007, 23, 62–68.
- Bauer, P.V.; Duca, F.A.; Waise, T.M.Z.; Rasmussen, B.A.; Abraham, M.A.; Dranse, H.J.; Puri, A.; O’Brien, C.A.; Lam, T.K.T. Metformin alters upper small intestinal microbiota that impact a glucose-sglt1-sensing glucoregulatory pathway. Cell Metab. 2018, 27, 101–117.e105.
- Rooj, A.K.; Kimura, Y.; Buddington, R.K. Metabolites produced by probiotic lactobacilli rapidly increase glucose uptake by caco-2 cells. BMC Microbiol. 2010, 10, 16.
- Fredborg, M.; Theil, P.K.; Jensen, B.B.; Purup, S. G protein-coupled receptor120 (gpr120) transcription in intestinal epithelial cells is significantly affected by bacteria belonging to the bacteroides, proteobacteria, and firmicutes phyla. J. Anim. Sci. 2012, 90 (Suppl. 4), 10–12.
- Tanaka, T.; Katsuma, S.; Adachi, T.; Koshimizu, T.A.; Hirasawa, A.; Tsujimoto, G. Free fatty acids induce cholecystokinin secretion through gpr120. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2008, 377, 523–527.
- Bauer, P.V.; Duca, F.A.; Waise, T.M.Z.; Dranse, H.J.; Rasmussen, B.A.; Puri, A.; Rasti, M.; O’Brien, C.A.; Lam, T.K.T. Lactobacillus gasseri in the upper small intestine impacts an acsl3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metab. 2018, 27, 572–587.e6.
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340.
- Karaki, S.; Tazoe, H.; Hayashi, H.; Kashiwabara, H.; Tooyama, K.; Suzuki, Y.; Kuwahara, A. Expression of the short-chain fatty acid receptor, gpr43, in the human colon. J. Mol. Histol. 2008, 39, 135–142.
- Karaki, S.; Mitsui, R.; Hayashi, H.; Kato, I.; Sugiya, H.; Iwanaga, T.; Furness, J.B.; Kuwahara, A. Short-chain fatty acid receptor, gpr43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006, 324, 353–360.
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the g-protein-coupled receptor ffar2. Diabetes 2012, 61, 364–371.
- Cherbut, C.; Ferrier, L.; Roze, C.; Anini, Y.; Blottiere, H.; Lecannu, G.; Galmiche, J.P. Short-chain fatty acids modify colonic motility through nerves and polypeptide yy release in the rat. Am. J. Physiol. 1998, 275, G1415–G1422.
- Holz, G.G., IV; Kuhtreiber, W.M.; Habener, J.F. Pancreatic beta-cells are rendered glucose-competent by the insulinotropic hormone glucagon-like peptide-1(7-37). Nature 1993, 361, 362–365.
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71.
- Lee, H.; Ko, G. Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol. 2014, 80, 5935–5943.
- Lee, H.; Lee, Y.; Kim, J.; An, J.; Lee, S.; Kong, H.; Song, Y.; Lee, C.K.; Kim, K. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 2018, 9, 155–165.
- Ryan, P.M.; Patterson, E.; Carafa, I.; Mandal, R.; Wishart, D.S.; Dinan, T.G.; Cryan, J.F.; Tuohy, K.M.; Stanton, C.; Ross, R.P. Metformin and dipeptidyl peptidase-4 inhibitor differentially modulate the intestinal microbiota and plasma metabolome of metabolically dysfunctional mice. Can. J. Diabetes 2020, 44, 146–155.e2.
- Zhang, W.; Xu, J.H.; Yu, T.; Chen, Q.K. Effects of berberine and metformin on intestinal inflammation and gut microbiome composition in db/db mice. Biomed. Pharmacother. 2019, 118, 109131.
- Ahmadi, S.; Razazan, A.; Nagpal, R.; Jain, S.; Wang, B.; Mishra, S.P.; Wang, S.; Justice, J.; Ding, J.; McClain, D.A.; et al. Metformin reduces aging-related leaky gut and improves cognitive function by beneficially modulating gut microbiome/goblet cell/mucin axis. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, e9–e21.
- Liu, G.; Liang, L.; Yu, G.; Li, Q. Pumpkin polysaccharide modifies the gut microbiota during alleviation of type 2 diabetes in rats. Int. J. Biol. Macromol. 2018, 115, 711–717.
- Liu, Y.; Wang, C.; Li, J.; Li, T.; Zhang, Y.; Liang, Y.; Mei, Y. Phellinus linteus polysaccharide extract improves insulin resistance by regulating gut microbiota composition. FASEB J. 2020, 34, 1065–1078.
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72.
- Rios-Covian, D.; Arboleya, S.; Hernandez-Barranco, A.M.; Alvarez-Buylla, J.R.; Ruas-Madiedo, P.; Gueimonde, M.; de los Reyes-Gavilan, C.G. Interactions between bifidobacterium and bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl. Environ. Microbiol. 2013, 79, 7518–7524.
- Hao, Z.; Li, L.; Ning, Z.; Zhang, X.; Mayne, J.; Cheng, K.; Walker, K.; Liu, H.; Figeys, D. Metaproteomics reveals growth phase-dependent responses of an in vitro gut microbiota to metformin. J. Am. Soc. Mass Spectrom. 2020, 31, 1448–1458.
- Pryor, R.; Norvaisas, P.; Marinos, G.; Best, L.; Thingholm, L.B.; Quintaneiro, L.M.; De Haes, W.; Esser, D.; Waschina, S.; Lujan, C.; et al. Host-microbe-drug-nutrient screen identifies bacterial effectors of metformin therapy. Cell 2019, 178, 1299–1312.e29.
- Brandt, A.; Hernandez-Arriaga, A.; Kehm, R.; Sanchez, V.; Jin, C.J.; Nier, A.; Baumann, A.; Camarinha-Silva, A.; Bergheim, I. Metformin attenuates the onset of non-alcoholic fatty liver disease and affects intestinal microbiota and barrier in small intestine. Sci. Rep. 2019, 9, 6668.
- Wankhade, U.D.; Zhong, Y.; Lazarenko, O.P.; Chintapalli, S.V.; Piccolo, B.D.; Chen, J.R.; Shankar, K. Sex-specific changes in gut microbiome composition following blueberry consumption in c57bl/6j mice. Nutrients 2019, 11, 313.
- Caslin, B.; Maguire, C.; Karmakar, A.; Mohler, K.; Wylie, D.; Melamed, E. Alcohol shifts gut microbial networks and ameliorates a murine model of neuroinflammation in a sex-specific pattern. Proc. Natl. Acad. Sci. USA 2019, 116, 25808–25815.
- Geer, E.B.; Shen, W. Gender differences in insulin resistance, body composition, and energy balance. Gend. Med. 2009, 6 (Suppl. 1), 60–75.
- Wu, B.N.; O’Sullivan, A.J. Sex differences in energy metabolism need to be considered with lifestyle modifications in humans. J. Nutr. Metab. 2011, 2011, 391809.
- Kornman, K.S.; Loesche, W.J. Effects of estradiol and progesterone on bacteroides melaninogenicus and bacteroides gingivalis. Infect. Immun. 1982, 35, 256–263.
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517.
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240.
- Li, X.; Wang, E.; Yin, B.; Fang, D.; Chen, P.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Effects of lactobacillus casei ccfm419 on insulin resistance and gut microbiota in type 2 diabetic mice. Benef. Microbes 2017, 8, 421–432.
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019, 26, 222–235.e5.
- Zheng, J.; Li, H.; Zhang, X.; Jiang, M.; Luo, C.; Lu, Z.; Xu, Z.; Shi, J. Prebiotic mannan-oligosaccharides augment the hypoglycemic effects of metformin in correlation with modulating gut microbiota. J. Agric. Food Chem. 2018, 66, 5821–5831.
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735.
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481.
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 2007, 50, 2374–2383.
- Tanti, J.F.; Gual, P.; Gremeaux, T.; Gonzalez, T.; Barres, R.; Le Marchand-Brustel, Y. Alteration in insulin action: Role of irs-1 serine phosphorylation in the retroregulation of insulin signalling. Ann. Endocrinol. 2004, 65, 43–48.
- Macchione, I.G.; Lopetuso, L.R.; Ianiro, G.; Napoli, M.; Gibiino, G.; Rizzatti, G.; Petito, V.; Gasbarrini, A.; Scaldaferri, F. Akkermansia muciniphila: Key player in metabolic and gastrointestinal disorders. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8075–8083.
- Kim, Y.S.; Ho, S.B. Intestinal goblet cells and mucins in health and disease: Recent insights and progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330.
- Johansson, M.E.; Larsson, J.M.; Hansson, G.C. The two mucus layers of colon are organized by the muc2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4659–4665.
- Amar, J.; Chabo, C.; Waget, A.; Klopp, P.; Vachoux, C.; Bermudez-Humaran, L.G.; Smirnova, N.; Berge, M.; Sulpice, T.; Lahtinen, S.; et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: Molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011, 3, 559–572.
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071.
- Belzer, C.; de Vos, W.M. Microbes inside—From diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458.
- Van Passel, M.W.; Kant, R.; Zoetendal, E.G.; Plugge, C.M.; Derrien, M.; Malfatti, S.A.; Chain, P.S.; Woyke, T.; Palva, A.; de Vos, W.M.; et al. The genome of akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 2011, 6, e16876.
- Ma, W.; Chen, J.; Meng, Y.; Yang, J.; Cui, Q.; Zhou, Y. Metformin alters gut microbiota of healthy mice: Implication for its potential role in gut microbiota homeostasis. Front. Microbiol. 2018, 9, 1336.
- Rosario, D.; Benfeitas, R.; Bidkhori, G.; Zhang, C.; Uhlen, M.; Shoaie, S.; Mardinoglu, A. Understanding the representative gut microbiota dysbiosis in metformin-treated type 2 diabetes patients using genome-scale metabolic modeling. Front. Physiol. 2018, 9, 775.
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405.
- Cui, H.X.; Zhang, L.S.; Luo, Y.; Yuan, K.; Huang, Z.Y.; Guo, Y. A purified anthraquinone-glycoside preparation from rhubarb ameliorates type 2 diabetes mellitus by modulating the gut microbiota and reducing inflammation. Front. Microbiol. 2019, 10, 1423.
- Ji, S.; Wang, L.; Li, L. Effect of metformin on short-term high-fat diet-induced weight gain and anxiety-like behavior and the gut microbiota. Front. Endocrinol. 2019, 10, 704.
- Chen, L.C.; Fan, Z.Y.; Wang, H.Y.; Wen, D.C.; Zhang, S.Y. Effect of polysaccharides from adlay seed on anti-diabetic and gut microbiota. Food Funct. 2019, 10, 4372–4380.
- Wang, J.H.; Bose, S.; Shin, N.R.; Chin, Y.W.; Choi, Y.H.; Kim, H. Pharmaceutical impact of houttuynia cordata and metformin combination on high-fat-diet-induced metabolic disorders: Link to intestinal microbiota and metabolic endotoxemia. Front. Endocrinol. 2018, 9, 620.
- Liu, G.; Bei, J.; Liang, L.; Yu, G.; Li, L.; Li, Q. Stachyose improves inflammation through modulating gut microbiota of high-fat diet/streptozotocin-induced type 2 diabetes in rats. Mol. Nutr. Food Res. 2018, 62, e1700954.
- Khat-Udomkiri, N.; Toejing, P.; Sirilun, S.; Chaiyasut, C.; Lailerd, N. Antihyperglycemic effect of rice husk derived xylooligosaccharides in high-fat diet and low-dose streptozotocin-induced type 2 diabetic rat model. Food Sci. Nutr. 2020, 8, 428–444.
- Wang, J.H.; Bose, S.; Lim, S.K.; Ansari, A.; Chin, Y.W.; Choi, H.S.; Kim, H. Houttuynia cordata facilitates metformin on ameliorating insulin resistance associated with gut microbiota alteration in oletf rats. Genes 2017, 8, 239.
- Zhang, M.; Feng, R.; Yang, M.; Qian, C.; Wang, Z.; Liu, W.; Ma, J. Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in zucker diabetic fatty rats. BMJ Open Diabetes Res. Care 2019, 7, e000717.
- Chelakkot, C.; Choi, Y.; Kim, D.K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.S.; Jee, Y.K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450.
- Derrien, M.; Collado, M.C.; Ben-Amor, K.; Salminen, S.; de Vos, W.M. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008, 74, 1646–1648.
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113.
- Artis, D.; Wang, M.L.; Keilbaugh, S.A.; He, W.; Brenes, M.; Swain, G.P.; Knight, P.A.; Donaldson, D.D.; Lazar, M.A.; Miller, H.R.; et al. Relmbeta/fizz2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc. Natl. Acad. Sci. USA 2004, 101, 13596–13600.
- Suemori, S.; Lynch-Devaney, K.; Podolsky, D.K. Identification and characterization of rat intestinal trefoil factor: Tissue- and cell-specific member of the trefoil protein family. Proc. Natl. Acad. Sci. USA 1991, 88, 11017–11021.
- Heilbronn, L.K.; Campbell, L.V. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr. Pharm. Des. 2008, 14, 1225–1230.
- Lumeng, C.N.; Saltiel, A.R. Inflammatory links between obesity and metabolic disease. J. Clin. Investig. 2011, 121, 2111–2117.
- Osborn, O.; Olefsky, J.M. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 2012, 18, 363–374.
- Brestoff, J.R.; Artis, D. Immune regulation of metabolic homeostasis in health and disease. Cell 2015, 161, 146–160.
- Lackey, D.E.; Olefsky, J.M. Regulation of metabolism by the innate immune system. Nat. Rev. Endocrinol. 2016, 12, 15–28.
- Lee, S.Y.; Lee, S.H.; Yang, E.J.; Kim, E.K.; Kim, J.K.; Shin, D.Y.; Cho, M.L. Metformin ameliorates inflammatory bowel disease by suppression of the stat3 signaling pathway and regulation of the between th17/treg balance. PLoS ONE 2015, 10, e0135858.
- Li, S.N.; Wang, X.; Zeng, Q.T.; Feng, Y.B.; Cheng, X.; Mao, X.B.; Wang, T.H.; Deng, H.P. Metformin inhibits nuclear factor kappab activation and decreases serum high-sensitivity c-reactive protein level in experimental atherogenesis of rabbits. Heart Vessel. 2009, 24, 446–453.
- Huang, N.L.; Chiang, S.H.; Hsueh, C.H.; Liang, Y.J.; Chen, Y.J.; Lai, L.P. Metformin inhibits tnf-alpha-induced ikappab kinase phosphorylation, ikappab-alpha degradation and il-6 production in endothelial cells through pi3k-dependent ampk phosphorylation. Int. J. Cardiol. 2009, 134, 169–175.
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in ibd mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428.
- Hansen, C.H.; Holm, T.L.; Krych, L.; Andresen, L.; Nielsen, D.S.; Rune, I.; Hansen, A.K.; Skov, S. Gut microbiota regulates nkg2d ligand expression on intestinal epithelial cells. Eur. J. Immunol. 2013, 43, 447–457.
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103.
- Rotter, V.; Nagaev, I.; Smith, U. Interleukin-6 (il-6) induces insulin resistance in 3t3-l1 adipocytes and is, like il-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J. Biol. Chem. 2003, 278, 45777–45784.
- Kang, C.S.; Ban, M.; Choi, E.J.; Moon, H.G.; Jeon, J.S.; Kim, D.K.; Park, S.K.; Jeon, S.G.; Roh, T.Y.; Myung, S.J.; et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 2013, 8, e76520.
- Weigert, C.; Hennige, A.M.; Brodbeck, K.; Haring, H.U.; Schleicher, E.D. Interleukin-6 acts as insulin sensitizer on glycogen synthesis in human skeletal muscle cells by phosphorylation of ser473 of akt. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E251–E257.
- Bharti, A.C.; Aggarwal, B.B. Nuclear factor-kappa b and cancer: Its role in prevention and therapy. Biochem. Pharmacol. 2002, 64, 883–888.
- Inan, M.S.; Rasoulpour, R.J.; Yin, L.; Hubbard, A.K.; Rosenberg, D.W.; Giardina, C. The luminal short-chain fatty acid butyrate modulates nf-kappab activity in a human colonic epithelial cell line. Gastroenterology 2000, 118, 724–734.
- Kinoshita, M.; Suzuki, Y.; Saito, Y. Butyrate reduces colonic paracellular permeability by enhancing ppargamma activation. Biochem. Biophys. Res. Commun. 2002, 293, 827–831.
- Li, X.; Wang, N.; Yin, B.; Fang, D.; Jiang, T.; Fang, S.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Effects of lactobacillus plantarum ccfm0236 on hyperglycaemia and insulin resistance in high-fat and streptozotocin-induced type 2 diabetic mice. J. Appl. Microbiol. 2016, 121, 1727–1736.
- Chen, P.; Zhang, Q.; Dang, H.; Liu, X.; Tian, F.; Zhao, J.; Chen, Y.; Zhang, H.; Chen, W. Antidiabetic effect of lactobacillus casei ccfm0412 on mice with type 2 diabetes induced by a high-fat diet and streptozotocin. Nutrition 2014, 30, 1061–1068.
- Tian, P.; Li, B.; He, C.; Song, W.; Hou, A.; Tian, S.; Meng, X.; Li, K.; Shan, Y. Antidiabetic (type 2) effects of lactobacillus g15 and q14 in rats through regulation of intestinal permeability and microbiota. Food Funct. 2016, 7, 3789–3797.
- Sun, K.Y.; Xu, D.H.; Xie, C.; Plummer, S.; Tang, J.; Yang, X.F.; Ji, X.H. Lactobacillus paracasei modulates lps-induced inflammatory cytokine release by monocyte-macrophages via the up-regulation of negative regulators of nf-kappab signaling in a tlr2-dependent manner. Cytokine 2017, 92, 1–11.
- Xue, J.; Li, X.; Liu, P.; Li, K.; Sha, L.; Yang, X.; Zhu, L.; Wang, Z.; Dong, Y.; Zhang, L.; et al. Inulin and metformin ameliorate polycystic ovary syndrome via anti-inflammation and modulating gut microbiota in mice. Endocr. J. 2019, 66, 859–870.
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983.
- Sansome, D.J.; Xie, C.; Veedfald, S.; Horowitz, M.; Rayner, C.K.; Wu, T. Mechanism of glucose-lowering by metformin in type 2 diabetes: Role of bile acids. Diabetes Obes. Metab. 2020, 22, 141–148.
- Caspary, W.F.; Creutzfeldt, W. Inhibition of bile salt absorption by blood-sugar lowering biguanides. Diabetologia 1975, 11, 113–117.
- Scarpello, J.H.; Hodgson, E.; Howlett, H.C. Effect of metformin on bile salt circulation and intestinal motility in type 2 diabetes mellitus. Diabet. Med. 1998, 15, 651–656.
- Meng, X.M.; Ma, X.X.; Tian, Y.L.; Jiang, Q.; Wang, L.L.; Shi, R.; Ding, L.; Pang, S.G. Metformin improves the glucose and lipid metabolism via influencing the level of serum total bile acids in rats with streptozotocin-induced type 2 diabetes mellitus. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2232–2237.
- Trabelsi, M.S.; Daoudi, M.; Prawitt, J.; Ducastel, S.; Touche, V.; Sayin, S.I.; Perino, A.; Brighton, C.A.; Sebti, Y.; Kluza, J.; et al. Farnesoid x receptor inhibits glucagon-like peptide-1 production by enteroendocrine l cells. Nat. Commun. 2015, 6, 7629.
- Li, F.; Jiang, C.; Krausz, K.W.; Li, Y.; Albert, I.; Hao, H.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid x receptor signalling and decreased obesity. Nat. Commun 2013, 4, 2384.
- Watanabe, M.; Horai, Y.; Houten, S.M.; Morimoto, K.; Sugizaki, T.; Arita, E.; Mataki, C.; Sato, H.; Tanigawara, Y.; Schoonjans, K.; et al. Lowering bile acid pool size with a synthetic farnesoid x receptor (fxr) agonist induces obesity and diabetes through reduced energy expenditure. J. Biol. Chem. 2011, 286, 26913–26920.
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring fxr antagonist. Cell Metab. 2013, 17, 225–235.
- Pathak, P.; Xie, C.; Nichols, R.G.; Ferrell, J.M.; Boehme, S.; Krausz, K.W.; Patterson, A.D.; Gonzalez, F.J.; Chiang, J.Y.L. Intestine farnesoid x receptor agonist and the gut microbiota activate g-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018, 68, 1574–1588.
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid x receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109.
- Cariou, B.; van Harmelen, K.; Duran-Sandoval, D.; van Dijk, T.H.; Grefhorst, A.; Abdelkarim, M.; Caron, S.; Torpier, G.; Fruchart, J.C.; Gonzalez, F.J.; et al. The farnesoid x receptor modulates adiposity and peripheral insulin sensitivity in mice. J. Biol. Chem. 2006, 281, 11039–11049.
- Zhang, Y.; Lee, F.Y.; Barrera, G.; Lee, H.; Vales, C.; Gonzalez, F.J.; Willson, T.M.; Edwards, P.A. Activation of the nuclear receptor fxr improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1006–1011.
- Clarke, S.F.; Murphy, E.F.; Nilaweera, K.; Ross, P.R.; Shanahan, F.; O’Toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes 2012, 3, 186–202.
- Louis, S.; Tappu, R.M.; Damms-Machado, A.; Huson, D.H.; Bischoff, S.C. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PLoS ONE 2016, 11, e0149564.
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014, 146, 1449–1458.
- Hu, S.; Wang, Y.; Lichtenstein, L.; Tao, Y.; Musch, M.W.; Jabri, B.; Antonopoulos, D.; Claud, E.C.; Chang, E.B. Regional differences in colonic mucosa-associated microbiota determine the physiological expression of host heat shock proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G1266–G1275.
- Harrell, L.; Wang, Y.; Antonopoulos, D.; Young, V.; Lichtenstein, L.; Huang, Y.; Hanauer, S.; Chang, E. Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS ONE 2012, 7, e32545.
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of e. Coli in the inflamed gut. Science 2013, 339, 708–711.
- Rhee, S.H. Lipopolysaccharide: Basic biochemistry, intracellular signaling, and physiological impacts in the gut. Intest. Res. 2014, 12, 90–95.
- Florez, H.; Pan, Q.; Ackermann, R.T.; Marrero, D.G.; Barrett-Connor, E.; Delahanty, L.; Kriska, A.; Saudek, C.D.; Goldberg, R.B.; Rubin, R.R.; et al. Impact of lifestyle intervention and metformin on health-related quality of life: The diabetes prevention program randomized trial. J. Gen. Intern. Med. 2012, 27, 1594–1601.
- Van Eldere, J.; Robben, J.; De Pauw, G.; Merckx, R.; Eyssen, H. Isolation and identification of intestinal steroid-desulfating bacteria from rats and humans. Appl. Environ. Microbiol. 1988, 54, 2112–2117.
- Manichanh, C.; Eck, A.; Varela, E.; Roca, J.; Clemente, J.C.; Gonzalez, A.; Knights, D.; Knight, R.; Estrella, S.; Hernandez, C.; et al. Anal gas evacuation and colonic microbiota in patients with flatulence: Effect of diet. Gut 2014, 63, 401–408.
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The seed and the rapid annotation of microbial genomes using subsystems technology (rast). Nucleic Acids Res. 2014, 42, D206–D214.
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016, 167, 1339–1353.e21.
- Roopchand, D.E.; Carmody, R.N.; Kuhn, P.; Moskal, K.; Rojas-Silva, P.; Turnbaugh, P.J.; Raskin, I. Dietary polyphenols promote growth of the gut bacterium akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 2015, 64, 2847–2858.
- Anhe, F.F.; Roy, D.; Pilon, G.; Dudonne, S.; Matamoros, S.; Varin, T.V.; Garofalo, C.; Moine, Q.; Desjardins, Y.; Levy, E.; et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015, 64, 872–883.
- Greer, R.L.; Dong, X.; Moraes, A.C.; Zielke, R.A.; Fernandes, G.R.; Peremyslova, E.; Vasquez-Perez, S.; Schoenborn, A.A.; Gomes, E.P.; Pereira, A.C.; et al. Akkermansia muciniphila mediates negative effects of ifngamma on glucose metabolism. Nat. Commun. 2016, 7, 13329.
- Zhang, H.; Sparks, J.B.; Karyala, S.V.; Settlage, R.; Luo, X.M. Host adaptive immunity alters gut microbiota. ISME J. 2015, 9, 770–781.
- Collado, M.C.; Derrien, M.; Isolauri, E.; de Vos, W.M.; Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770.
- Kong, F.; Hua, Y.; Zeng, B.; Ning, R.; Li, Y.; Zhao, J. Gut microbiota signatures of longevity. Curr. Biol. 2016, 26, R832–R833.
- Gupta, V.K.; Kim, M.; Bakshi, U.; Cunningham, K.Y.; Davis, J.M., 3rd; Lazaridis, K.N.; Nelson, H.; Chia, N.; Sung, J. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 2020, 11, 4635.
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16s rrna gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029.
- Yang, J.; Pu, J.; Lu, S.; Bai, X.; Wu, Y.; Jin, D.; Cheng, Y.; Zhang, G.; Zhu, W.; Luo, X.; et al. Species-level analysis of human gut microbiota with metataxonomics. Front. Microbiol. 2020, 11, 2029.
