Coronaviruses (CoVs) are positive-sense RNA enveloped viruses, members of the family Coronaviridae, that cause infections in a broad range of mammals including humans. Several CoV species lead to mild upper respiratory infections typically associated with common colds. However, three human CoV (HCoV) species: Severe Acute Respiratory Syndrome (SARS)-CoV-1, Middle East Respiratory Syndrome (MERS)-CoV, and SARS-CoV-2, are responsible for severe respiratory diseases at the origin of two recent epidemics (SARS and MERS), and of the current COronaVIrus Disease 19 (COVID-19), respectively.
- coronavirus
- SARS-CoV-1
- SARS-CoV-2
- MERS-CoV
- nucleoside
- remdesivir
- lopinavir
- ritonavir
- favipiravir
1. Introduction
Coronaviruses (CoVs) are enveloped positive-sense single-stranded RNA viruses belonging to the family Coronaviridae, causing infections in avian species, mammals, and, among these, humans
. Human coronaviruses (HCoV) are believed to be of zoonotic origin, and their infections mainly lead to respiratory diseases
. In particular, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 cause the mild seasonal symptoms of the common cold
. However, three HCoV species responsible for the onset of life-threatening respiratory events emerged in the last two decades: Severe Acute Respiratory Syndrome (SARS)-CoV-1, Middle East Respiratory Syndrome (MERS)-CoV, and SARS-CoV-2
. Human infection by SARS-CoV-2 is at the origin of the current COronaVIrus Disease 19 (COVID-19) pandemic. Interestingly, SARS-CoV-1 and MERS-CoV are more lethal but less transmissible than SARS-CoV-2, to which they are closely related
. There is clearly an urgent need for mass immunization and specific treatments for these HCoV-associated pathologies. CoV infection starts with the specific molecular recognition between the CoV spike (S) protein and host-specific receptors exposed on the surface of the target cells
. These have been identified for several CoVs and represent the primary molecular targets for anti-CoV strategies
. Human aminopeptidase N (APN) is involved in the infection by HCoV-229E; 9-O-acetylated sialic acid (9-O-Ac-Sia) receptor for HCoV-OC43 and HCoV-HKU1; angiotensin-converting enzyme 2 (ACE2) for HCoV-NL63, SARS-CoV-1, and SARS-CoV-2; dipeptidyl peptidase 4 (DPP4) for MERS-CoV
. Intracellularly, CoVs replicate their RNA and produce the viral proteins required for the assembly of new viral particles
. While five out of the seven HCoVs are usually associated with mild upper respiratory infections, MERS-CoV and SARS-CoV-1 and 2 can lead to lethal events
. In particular, the new SARS-CoV-2, first emerging in China at the end of 2019
, can provoke severe pneumonia, and being easily transmissible, it rapidly spread worldwide leading the World Health Organization (WHO) to declare COVID-19 a pandemic in March 2020
. Currently, there have been more than two million deaths due to COVID-19 (2,239,418 as found in Worldometers.info
accessed on 1 February 2021), with enormous consequences for public health and the global economy
. While the whole world is fighting against COVID-19 and waits for a global and effective vaccination, the scientific community is devoting immense efforts to develop effective drugs for the immediate treatment of SARS-CoV-2 infection. Due to the urgent need for such a pharmacological treatment, drug repurposing
is one of the most common approaches. In this context, nucleobase-containing synthetic molecules
and modified nucleosides
are attracting significant interest for their antiviral activity
. In particular, nucleoside-mimicking analogs
, as well as nucleoside precursors
, being able to inhibit the growth of viruses, play a pivotal role in the search of effective therapies for HCoV infectious diseases
.
2. Human Coronaviruses
Presently, seven HCoVs are known and described in the scientific literature
. Besides the well-known potentially lethal SARS-CoV-1, MERS-CoV, and SARS-CoV-2, the common human coronaviruses HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 identified in the last few decades were classified into two CoV genera: Alphacoronavirus and Betacoronavirus
.
HCoV-229E and HCoV-NL63, belonging to the genus Alphacoronavirus
, are genetically related to each other and are responsible for about 5% of all respiratory infections in hospitalized children
. Both HCoV-OC43 and HCoV-HKU1, of the genus Betacoronavirus
, are ‘common cold’ viruses widely circulating worldwide, with associated severity of respiratory symptoms being documented only in rare cases
. Even though these HCoVs do not cause severe clinical symptoms in most patients, HCoV 229E and OC43 can provoke pneumonia
, while HCoV-NL63 and HCoV-HKU1 infection lead in some cases also to bronchiolitis and croup
.
3. Prophylaxis and Therapy of HCoV Diseases
Before 2002, the only known HCoVs were the common human coronaviruses associated with ‘common cold’ symptoms mentioned above. Since most people with an illness caused by them usually recovered spontaneously, there was typically no need for any drugs other than aspirin to relieve the cold-associated symptoms. Conversely, the critical clinical conditions often observed in patients affected by the highly pathogenic MERS-CoV, SARS-CoV-1, and, especially, SARS-CoV-2 recalled the urgency of developing vaccines and antiviral treatments for HCoV infections. This research theme was previously almost ignored by pharmaceutical companies and, in our opinion, should not be abandoned by the scientific community even when the COVID-19 emergency will be over. In SARS-CoV-1 infection, scientists undertook initial vaccine studies, but the obtained candidates presented severe complications such as immune disease insurgence in treated animals
. The research for other SARS vaccines was discontinued not only for the difficulties encountered, but mainly because SARS-CoV-1 vanished
. Owing to the pharmacological strategies adopted by physicians for SARS patients, these were essentially empirical and involved repurposed immunomodulatory and antiviral drugs, such as corticosteroids, lopinavir/ritonavir, and ribavirin
. Concerning MERS, despite several efforts to search for effective vaccines, antibodies, and drugs, no conclusive results were achieved. Repurposed drugs used with some success for MERS include again lopinavir/ritonavir and ribavirin
. After this premise, it appears clear how the lack of any useful vaccine and drug against SARS-CoV-1 and MERS-CoV was reflected in the current crisis, considering the relatively close relationship between SARS-CoV-1 and SARS-CoV-2 genomes, as well as the conserved nature of MERS and SARS-CoV-2 proteins
. Fortunately, academic institutions and pharmaceutical companies have lately developed some promising vaccine candidates against SARS-CoV-2. Among them, the Pfizer-BioNTech (BNT162b2)
, the Moderna (mRNA-1273)
, and the Oxford University/AstraZeneca (ChAdOx1-S)
vaccines were authorized for prophylaxis of COVID-19, while several others are currently in late-stage clinical testing
. Vaccines may represent a medium/long-term solution to the current pandemic, but short-term solutions such as pharmacological treatments against SARS-CoV-2 remain urgently needed. Presently, SARS-CoV-2 infection therapy includes immunomodulatory drugs, plasma from individuals recovered from COVID-19, and several pharmacological treatments
. In this regard, despite numerous repurposed drugs being tested, only the nucleoside analog remdesivir has been officially approved by the American Food and Drug Administration (FDA) agency to date
.
4. Protein Targets for Anti-HCoV Pharmaceutical Strategies
Amongst the coronavirus targets that were studied, or are currently being investigated, in the fight against the three most pathogenic HCoVs, particular relevance is given to the spike (S) protein
, RNA-dependent RNA-polymerase (RdRp)
, papain-like protease (PL
)
and main protease (M
, 3CL
)
. In particular, this latter, which proteolytically cleaves the polyproteins to functional proteins essential for viral replication, occupies a special place in pharmaceutical research
. Hence, the frequently-reported administration of potential M
inhibitors like lopinavir and ritonavir to SARS, MERS, and COVID patients
, even though there is no agreement on the real efficacy of this cocktail therapy, especially in the case of the current pandemic
. RdRp is a protein involved in SARS-CoV-2 replication, considered to be conserved within RNA viruses
. Targeting the RdRp by antiviral drugs could be a potential therapeutic option to inhibit coronavirus RNA polymerization and, consequently, viral replication. Since remdesivir
, the only FDA-approved drug for COVID-19 available to date, is believed to inhibit SARS-CoV-2 RNA polymerase competing with natural nucleotide triphosphates for incorporation into growing viral RNA, this aspect attracts interest not only on this drug but also on other analogs of nucleosides and nucleoside precursors with similar RdRp inhibitory activity.
5. Synthetic Nucleoside Precursors for HCoV Disease Therapy
Among the synthetic drugs under scrutiny in the treatment of viral respiratory pathologies, nucleoside precursors occupy an important place, especially in the present COVID-19 pandemic
. Here below, we report on the main nucleoside precursors evaluated as anti-HCoV drugs (
).
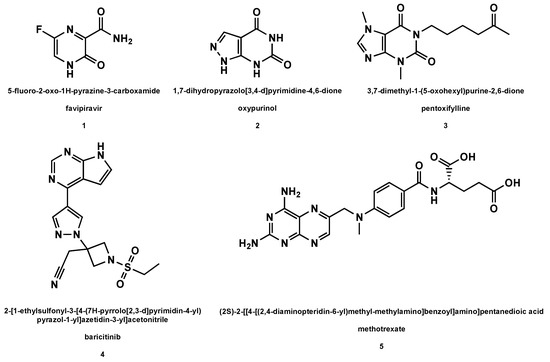
Structures of the nucleoside precursors and derivatives of purine analogs (with drug names and related IUPAC nomenclature) used or under investigation in the HCoV disease therapy.
References
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020.
- Chu, D.K.W.; Leung, C.Y.H.; Gilbert, M.; Joyner, P.H.; Ng, E.M.; Tse, T.M.; Guan, Y.; Peiris, J.S.M.; Poon, L.L.M. Avian Coronavirus in Wild Aquatic Birds. J. Virol. 2011, 85, 12815–12820.
- Poon, L.L.M.; Chu, D.K.W.; Chan, K.H.; Wong, O.K.; Ellis, T.M.; Leung, Y.H.C.; Lau, S.K.P.; Woo, P.C.Y.; Suen, K.Y.; Yuen, K.Y.; et al. Identification of a Novel Coronavirus in Bats. J. Virol. 2005, 79, 2001–2009.
- Wang, L.-F.; Anderson, D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019, 34, 79–89.
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; AlHakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823.
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pohlmann, S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993.
- Gossner, C.; Danielson, N.; Gervelmeyer, A.; Berthe, F.; Faye, B.; Kaasik Aaslav, K.; Adlhoch, C.; Zeller, H.; Penttinen, P.; Coulombier, D. Human–Dromedary Camel Interactions and the Risk of Acquiring Zoonotic Middle East Respiratory Syndrome Coronavirus Infection. Zoonoses Public Health 2014, 63, 1–9.
- Sheahan, T.; Rockx, B.; Donaldson, E.; Sims, A.; Pickles, R.; Corti, D.; Baric, R. Mechanisms of Zoonotic Severe Acute Respiratory Syndrome Coronavirus Host Range Expansion in Human Airway Epithelium. J. Virol. 2008, 82, 2274–2285.
- Sheahan, T.; Rockx, B.; Donaldson, E.; Corti, D.; Baric, R. Pathways of Cross-Species Transmission of Synthetically Reconstructed Zoonotic Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2008, 82, 8721–8732.
- Gaunt, E.R.; Hardie, A.; Claas, E.C.J.; Simmonds, P.; Templeton, K.E. Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method. J. Clin. Microbiol. 2010, 48, 2940–2947.
- Woldemeskel, B.A.; Kwaa, A.K.; Garliss, C.C.; Laeyendecker, O.; Ray, S.C.; Blankson, J.N. Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J. Clin. Investig. 2020, 130, 6631–6638.
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV-1 and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding and Infectiousness: A Systematic Review and Meta-Analysis. Lancet Microbe 2020, 2, e13–e22.
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452.
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184.
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726.
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403.
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pohlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2013, 88, 1293–1307.
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033.
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57.
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121.
- Pillay, T.S. Gene of the month: The 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020, 73, 366–369.
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355.
- Kim, C.-H. SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus–Host Interaction. Int. J. Mol. Sci. 2020, 21, 4549.
- Artese, A.; Svicher, V.; Costa, G.; Salpini, R.; Di Maio, V.C.; Alkhatib, M.; Ambrosio, F.A.; Santoro, M.M.; Assaraf, Y.G.; Alcaro, S.; et al. Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses. Drug Resist. Updates 2020, 53, 100721.
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol Biol. 2015, 1282, 1–23.
- Chen, B.; Tian, E.-K.; He, B.; Tian, L.; Han, R.; Wang, S.; Xiang, Q.; Zhang, S.; El Arnaout, T.; Cheng, W. Overview of lethal human coronaviruses. Signal Transduct. Target. 2020, 5, 1–16.
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160.
- Mercatelli, D.; Holding, A.N.; Giorgi, F.M. Web tools to fight pandemics: The COVID-19 experience. Brief. Bioinform. 2020.
- Arthi, V.; Parman, J. Disease, downturns, and wellbeing: Economic history and the long-run impacts of COVID-19. Explor. Econ. Hist. 2020, 79, 101381–101400.
- Roviello, V.; Roviello, G.N. Lower COVID-19 mortality in Italian forested areas suggests immunoprotection by Mediterranean plants. Env. Chem. Lett. 2020, 19, 699–710.
- Ibn-Mohammed, T.; Mustapha, K.B.; Godsell, J.; Adamu, Z.; Babatunde, K.A.; Akintade, D.D.; Acquaye, A.; Fujii, H.; Ndiaye, M.M.; Yamoah, F.A.; et al. A critical analysis of the impacts of COVID-19 on the global economy and ecosystems and opportunities for circular economy strategies. Resour. Conserv. Recycl. 2021, 164, 105169.
- Costanzo, M.; De Giglio, M.A.R.; Roviello, G.N. SARS CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and Other Drugs for the Treatment of the New Coronavirus. Curr. Med. Chem. 2020, 27, 4536–4541.
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharm. Rep. 2020, 72, 1479–1508.
- Musumeci, D.; Roviello, V.; Roviello, G.N. DNA- and RNA-binding ability of oligoDapT, a nucleobase-decorated peptide, for biomedical applications. Int. J. Nanomed. 2018, 13, 2613–2629.
- Roviello, G.N.; Vicidomini, C.; Costanzo, V.; Roviello, V. Nucleic acid binding and other biomedical properties of artificial oligolysines. Int. J. Nanomed. 2016, 11, 5897–5904.
- Roviello, G.N. Novel insights into nucleoamino acids: Biomolecular recognition and aggregation studies of a thymine-conjugated l-phenyl alanine. Amino Acids 2018, 50, 933–941.
- Musumeci, D.; Mokhir, A.; Roviello, G.N. Synthesis and nucleic acid binding evaluation of a thyminyl L-diaminobutanoic acid-based nucleopeptide. Bioorg. Chem. 2020, 100, 103862.
- Roviello, G.N.; Benedetti, E.; Pedone, C.; Bucci, E.M. Nucleobase-containing peptides: An overview of their characteristic features and applications. Amino Acids 2010, 39, 45–57.
- Roviello, G.N.; Musumeci, D. Synthetic approaches to nucleopeptides containing all four nucleobases, and nucleic acid-binding studies on a mixed-sequence nucleo-oligolysine. RSC Adv 2016, 6, 63578–63585.
- Roviello, G.N.; Musumeci, D.; Moccia, M.; Castiglione, M.; Sapio, R.; Valente, M.; Bucci, E.M.; Perretta, G.; Pedone, C. dabPNA: Design, synthesis, and DNA binding studies. Ncleosides Nucleotides Nucleic Acids 2007, 26, 1307–1310.
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Piccialli, G.; Bucci, E.; Piccialli, V.; Mayol, L. Synthesis of 4-N-alkyl and ribose-modified AICAR analogs on solid support. Tetrahedron 2008, 64, 6475–6481.
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Piccialli, G.; Mayol, L. Synthesis of N-1 and ribose modified inosine analogs on solid support. Tetrahedron Lett. 2007, 48, 397–400.
- Oliviero, G.; D’Errico, S.; Borbone, N.; Amato, J.; Piccialli, V.; Varra, M.; Piccialli, G.; Mayol, L. A solid-phase approach to the synthesis of N-1-alkyl analogs of cyclic inosine-diphosphate-ribose (cIDPR). Tetrahedron 2010, 66, 1931–1936.
- D’Errico, S.; Oliviero, G.; Amato, J.; Borbone, N.; Cerullo, V.; Hemminki, A.; Piccialli, V.; Zaccaria, S.; Mayol, L.; Piccialli, G. Synthesis and biological evaluation of unprecedented ring-expanded nucleosides (RENs) containing the imidazo[4,5-d][1,2,6]oxadiazepine ring system. Chem. Commun. 2012, 48, 9310.
- Oliviero, G.; D’Errico, S.; Borbone, N.; Amato, J.; Piccialli, V.; Piccialli, G.; Mayol, L. Facile Solid-Phase Synthesis of AICAR 5′-Monophosphate (ZMP) and Its 4-N-Alkyl Derivatives. Eur. J. Org. Chem. 2010, 2010, 1517–1524.
- D’Errico, S.; Oliviero, G.; Borbone, N.; Amato, J.; D’Alonzo, D.; Piccialli, V.; Mayol, L.; Piccialli, G. A Facile Synthesis of 5′-Fluoro-5′-deoxyacadesine (5′-F-AICAR): A Novel Non-phosphorylable AICAR Analog. Molecules 2012, 17, 13036–13044.
- Roviello, G.N.; Gaetano, S.D.; Capasso, D.; Cesarani, A.; Bucci, E.M.; Pedone, C. Synthesis, spectroscopic studies and biological activity of a novel nucleopeptide with Moloney murine leukemia virus reverse transcriptase inhibitory activity. Amino Acids 2009, 38, 1489–1496.
- Roviello, G.N.; Di Gaetano, S.; Capasso, D.; Franco, S.; Crescenzo, C.; Bucci, E.M.; Pedone, C. RNA-Binding and Viral Reverse Transcriptase Inhibitory Activity of a Novel Cationic Diamino Acid-Based Peptide. J. Med. Chem. 2011, 54, 2095–2101.
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analog antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir. Res. 2018, 154, 66–86.
- Eyer, L.; Nencka, R.; de Clercq, E.; Seley-Radtke, K.; Růžek, D. Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antivir. Chem. Chemother. 2018, 26, 204020661876129.
- Menéndez-Arias, L.; Jin, Z.; Smith, L.K.; Rajwanshi, V.K.; Kim, B.; Deval, J. The Ambiguous Base-Pairing and High Substrate Efficiency of T-705 (Favipiravir) Ribofuranosyl 5′-Triphosphate towards Influenza A Virus Polymerase. PLoS ONE 2013, 8, e68347.
- Jin, Z.; Tucker, K.; Lin, X.; Kao, C.C.; Shaw, K.; Tan, H.; Symons, J.; Behera, I.; Rajwanshi, V.K.; Dyatkina, N.; et al. Biochemical Evaluation of the Inhibition Properties of Favipiravir and 2′-C-Methyl-Cytidine Triphosphates against Human and Mouse Norovirus RNA Polymerases. Antimicrob. Agents Chemother. 2015, 59, 7504–7516.
- Boretti, A. Favipiravir use for SARS CoV-2 infection. Pharm. Rep. 2020, 72, 1542–1552.
- Kaptein, S.J.F.; Jacobs, S.; Langendries, L.; Seldeslachts, L.; ter Horst, S.; Liesenborghs, L.; Hens, B.; Vergote, V.; Heylen, E.; Barthelemy, K.; et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2−infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. USA 2020, 117, 26955–26965.
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697.
- Palù, G.; Parolin, C.; Calistri, A.; Salata, C. Coronaviruses: A paradigm of new emerging zoonotic diseases. Pathog. Dis. 2020, 77, ftaa006.
- Dijkman, R.; Jebbink, M.F.; El Idrissi, N.B.; Pyrc, K.; Muller, M.A.; Kuijpers, T.W.; Zaaijer, H.L.; van der Hoek, L. Human Coronavirus NL63 and 229E Seroconversion in Children. J. Clin. Microbiol. 2008, 46, 2368–2373.
- Sastre, P.; Dijkman, R.; Camuñas, A.; Ruiz, T.; Jebbink, M.F.; van der Hoek, L.; Vela, C.; Rueda, P. Differentiation between Human Coronaviruses NL63 and 229E Using a Novel Double-Antibody Sandwich Enzyme-Linked Immunosorbent Assay Based on Specific Monoclonal Antibodies. Clin. Vaccine Immunol. 2011, 18, 113–118.
- Al-Khannaq, M.N.; Ng, K.T.; Oong, X.Y.; Pang, Y.K.; Takebe, Y.; Chook, J.B.; Hanafi, N.S.; Kamarulzaman, A.; Tee, K.K. Molecular epidemiology and evolutionary histories of human coronavirus OC43 and HKU1 among patients with upper respiratory tract infections in Kuala Lumpur, Malaysia. Virol. J. 2016, 13, 1–12.
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Ref. Mod. Life Sci. 2020.
- Pene, F.; Merlat, A.; Vabret, A.; Rozenberg, F.; Buzyn, A.; Dreyfus, F.; Cariou, A.; Freymuth, F.; Lebon, P. Coronavirus 229E-Related Pneumonia in Immunocompromised Patients. Clin. Infect. Dis. 2003, 37, 929–932.
- Jordan, P.C.; Stevens, S.K.; Deval, J. Nucleosides for the treatment of respiratory RNA virus infections. Antivir. Chem. Chemother. 2018, 26, 204020661876448.
- Abdul-Rasool, S.; Fielding, B.C. Understanding Human Coronavirus HCoV-NL63. Open Virol. J. 2010, 4, 76–84.
- Esper, F.; Weibel, C.; Ferguson, D.; Landry, M.L.; Kahn, J.S. Coronavirus HKU1 Infection in the United States. Emerg. Infect. Dis. 2006, 12, 775–779.
- Hung, L.S. The SARS epidemic in Hong Kong: What lessons have we learned? JRSM 2003, 96, 374–378.
- Centers for Disease Control and Prevention. Outbreak of severe acute respiratory syndrome—Worldwide, 2003. MMWR Morb. Mortal. Wkly. Rep. 2003, 52, 226–228.
- Centers for Disease Control and Prevention. In the Absence of SARS-CoV Transmission Worldwide: Guidance for Surveillance, Clinical and Laboratory Evaluation, and Reporting; Version 2; CDC: Atlanta, GA, USA, 2005.
- World Health Organization. Middle East Respiratory Syndrome Coronavirus (MERS-CoV); WHO: Geneva, Switzerland, 2019.
- Oboho, I.K.; Tomczyk, S.M.; Al-Asmari, A.M.; Banjar, A.A.; Al-Mugti, H.; Aloraini, M.S.; Alkhaldi, K.Z.; Almohammadi, E.L.; Alraddadi, B.M.; Gerber, S.I.; et al. 2014 MERS-CoV Outbreak in Jeddah—A Link to Health Care Facilities. N. Engl. J. Med. 2015, 372, 846–854.
- Yi, Y.; Lagniton, P.N.P.; Ye, S.; Li, E.; Xu, R.-H. COVID-19: What has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020, 16, 1753–1766.
- Sallard, E.; Halloy, J.; Casane, D.; van Helden, J.; Decroly, E. Tracing the origins of SARS-COV-2 in coronavirus phylogenies. Med. Sci (Paris) 2020, 36, 783–796.
- Lvov, D.K.; Alkhovsky, S.V. Source of the COVID-19 pandemic: Ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus). Probl. Virol. Russ. J. 2020, 65, 62–70.
- Gaye, B.; Fanidi, A.; Jouven, X. Denominator matters in estimating COVID-19 mortality rates. Eur. Heart J. 2020, 41, 3500.
- Bolles, M.; Deming, D.; Long, K.; Agnihothram, S.; Whitmore, A.; Ferris, M.; Funkhouser, W.; Gralinski, L.; Totura, A.; Heise, M.; et al. A Double-Inactivated Severe Acute Respiratory Syndrome Coronavirus Vaccine Provides Incomplete Protection in Mice and Induces Increased Eosinophilic Proinflammatory Pulmonary Response upon Challenge. J. Virol. 2011, 85, 12201–12215.
- Khalili, J.S.; Zhu, H.; Mak, N.S.A.; Yan, Y.; Zhu, Y. Novel coronavirus treatment with ribavirin: Groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020, 92, 740–746.
- Chan, J.F.-W.; Yao, Y.; Yeung, M.-L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment with Lopinavir/Ritonavir or Interferon-β1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015, 212, 1904–1913.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020.
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2020, 397, 99–111.
- Parker, E.P.; Shrotri, M.; Kampmann, B. Keeping track of the SARS-CoV-2 vaccine pipeline. Nat. Rev. Immunol. 2020, 20, 650.
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020.
- Twomey, J.D.; Luo, S.; Dean, A.Q.; Bozza, W.P.; Nalli, A.; Zhang, B. COVID-19 update: The race to therapeutic development. Drug Resist. Updates 2020, 53, 100733.
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782.
- Zhu, W.; Chen, C.Z.; Gorshkov, K.; Xu, M.; Lo, D.C.; Zheng, W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. Slas Discov. Adv. Sci. Drug Discov. 2020, 25, 1141–1151.
- Imbert, I.; Guillemot, J.C.; Bourhis, J.M.; Bussetta, C.; Coutard, B.; Egloff, M.P.; Ferron, F.; Gorbalenya, A.E.; Canard, B. A second, non-canonical RNA-dependent RNA polymerase in SARS Coronavirus. EMBO J. 2006, 25, 4933–4942.
- Min, J.S.; Kim, G.-W.; Kwon, S.; Jin, Y.-H. A Cell-Based Reporter Assay for Screening Inhibitors of MERS Coronavirus RNA-Dependent RNA Polymerase Activity. J. Clin. Med. 2020, 9, 2399.
- Lu, A.; Zhang, H.; Zhang, X.; Wang, H.; Hu, Q.; Shen, L.; Schaffhausen, B.S.; Hou, W.; Li, L. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology 2004, 324, 84–89.
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 2014, 450, 64–70.
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275.
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335.
- Hilgenfeld, R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. 2014, 281, 4085–4096.
- Sharma, P.; Vijayan, V.; Pant, P.; Sharma, M.; Vikram, N.; Kaur, P.; Singh, T.; Sharma, S. Identification of potential drug candidates to combat COVID-19: A structural study using the main protease (mpro) of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 1–11.
- Chu, C.; Cheng, V.; Hung, I.; Wong, M.; Chan, K.; Chan, K.; Kao, R.; Poon, L.; Wong, C.; Guan, Y. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256.
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020.
- Wang, Y.; Anirudhan, V.; Du, R.; Cui, Q.; Rong, L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2020.
