Eukaryotic cells, are complex cells that evolved through endosymbiosis when one cell (typically bacterial, forming mitochondria and plastids) is incorporated by a host cell. It might well be that other cellular organelles are also of endosymbiotic nature.
- actin
- cell
- cell biology
- cytoskeleton
- consciousness
- eukaryotes
- excitability
- membranes
- sentience
- symbiosis
1. Chimeric Nature of the Eukaryotic Cell
[3]
[6]
[13]
2. Structures and Processes Behind Cellular Consciousness—Evolution of Chimeric Consciousness of Eukaryotic Cell
[8]
Box 1.

Box 2.

Vibrations of excitable polymers contribute to the intracellular electromagnetic fields and can be expected to interact with the field emanating from the excitable plasma membrane. As microtubules act as memristors, as combinations of memory and electromagnetic resistance [31], they are well suited to faithfully decode the cellular senomic fields and to act accordingly. Furthermore, microtubules are structurally linked to both the actin filaments as well as the plasma membrane; they are perfectly suited to generate subcellular bioelectric circuits [31][32].
3. Structures and Processes behind Cellular Consciousness—Two Types of Nanobrains Generating Consciousness of Eukaryotic Cell
Two ancient cells merging into one resulted in the generation of supracellular chimeric consciousness having four different excitable sources: two plasma membranes, F-actin, and microtubules. The plasma membrane of the host cells, associated with the actin cytoskeleton, produced the senomic fields of contemporary chimeric eukaryotic cells. The guest cell transformed into the eukaryotic nucleus with the centrosome associated with centriole and organizing perinuclear microtubules [33]. The plasma membrane and the nuclear envelope/centrosome/microtubules complex can be viewed as two different cellular nanobrains, the origin, of which can be traced back to the two ancient cells, which merged together, forming the first eukaryotic cell [4][5][6]. Vibrations of F-actin and microtubules contribute significantly to the cellular electromagnetic field [27][28]. As microtubules act both as intracellular nanowires and memristors, they are perfectly suited for the nanobrain roles of the centrosomes/nuclear envelopes, complementing the principle nanobrain of the eukaryotic cell represented by the excitable plasma membrane (Figure 1, Box 1 and Box 2) inherently linked to the actin cytoskeleton.
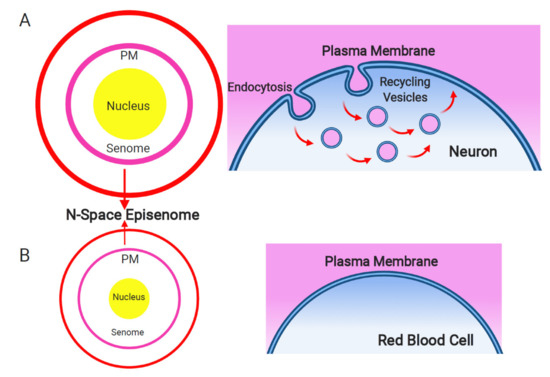
Figure 1. Plasma membrane and endosomal recycling vesicles-based nanobrain. Schematic depiction of the senome and the N-space episenome in two contrasting cells of multicellular organisms. (A) In the neurons and neuron-like cells, highly active endocytosis and endocytic vesicle recycling results in hypertrophied senome (lilac circle) and N-space episenome (red circle). Such cells are well-informed about their environment and are active in cell–cell communication via their plasma membrane-based nanobrains. (B) In the example of mature red blood cells, there are only minimal activities of endocytosis and endosomal vesicle recycling. Such cells have shrunk their senomes (lilac circle) and N-space episenomes (red circle) based nanobrains. They are socially isolated, with minimal cell–cell communication and highly reduced cellular sensory apparatus.
References
- Sarai, C.; Tanifuji, G.; Nakayama, T.; Kamikawa, R.; Takahashi, K.; Yazaki, E.; Matsuo, E.; Miyashita, H.; Ishida, K.I.; Iwataki, M.; et al. Dinoflagellates with relic endosymbiont nuclei as models for elucidating organellogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 5364–5375.
- Nakayama, T.; Takahashi, K.; Kamikawa, R.; Iwataki, M.; Inagaki, Y.; Tanifuji, G. Putative genome features of relic green alga-derived nuclei in dinoflagellates and future perspectives as model organisms. Commun. Integr. Biol. 2020, 13, 84–88.
- Sibbald, S.J.; Archibald, J.M. Genomic insights into plastid evolution. Genome Biol. Evol. 2020, 12, 978–990.
- Baluška, F.; Lyons, S. Energide-cell body as smallest unit of eukaryotic life. Ann. Bot. 2018, 122, 741–745.
- Baluška, F.; Lyons, S. Symbiotic origin of eukaryotic nucleus—From cell body to Neo-Energide. In Concepts in Cell Biology—History and Evolution; Sahi, V., Baluška, F., Eds.; Plant Cell Monographs 23; Springer International Publishing: Cham, Switzerland, 2018; pp. 39–66.
- Baluška, F.; Lyons, S. Archaeal origins of eukaryotic cell and nucleus. Biosystems 2021, 203, 104375.
- Margulis, L. Serial endosymbiotic theory (SET) and composite individuality. Transition from bacterial to eukaryotic genomes. Microbiol. Today 2004, 31, 172–174.
- Margulis, L. The conscious cell. Ann. N Y Acad. Sci. 2001, 929, 55–70.
- De Duve, C. The origin of eukaryotes: A reappraisal. Nat. Rev. Genet. 2007, 8, 395–403.
- Zachar, I.; Boza, G. Endosymbiosis before eukaryotes: Mitochondrial establishment in protoeukaryotes. Cell. Mol. Life Sci. 2020, 77, 3503–3523.
- Patron, N.J.; Waller, R.F.; Keeling, P.J. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 2006, 357, 1373–1382.
- Hehenberger, E.; Gast, R.J.; Keeling, P.J. A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc. Natl. Acad. Sci. USA 2019, 116, 17934–17942.
- Jacob, F. Evolution and tinkering. Science 1977, 196, 1161–1166.
- Agnati, L.F.; Baluška, F.; Barlow, P.W.; Guidolin, D. Mosaic, self-similarity logic, and biological attraction principles: Three explanatory instruments in biology. Commun. Integr. Biol. 2009, 2, 552–563.
- Agnati, L.F.; Barlow, P.W.; Baldelli, E.; Baluška, F. Are maternal mitochondria the selfish entities that are masters of the cells of eukaryotic multicellular organisms? Commun. Integr. Biol. 2009, 2, 194–200.
- Baluška, F.; Volkmann, D.; Barlow, P.W. Eukaryotic cells and their Cell Bodies: Cell Theory revisited. Ann. Bot. 2004, 94, 9–32.
- Baluška, F.; Reber, A. Sentience and consciousness in single cells: How the first minds emerged in unicellular species. BioEssays 2019, 41, e1800229.
- Reber, A.S.; Baluška, F. Cognition is some surprising places. Biochem. Biophys. Res. Commun. 2021, in press.
- Baluška, F.; Miller, W.B., Jr. Senomic view of the cell: Senome versus Genome. Commun. Integr. Biol. 2018, 11, 1–9.
- Miller, W.B., Jr.; Torday, J.S.; Baluška, F. The N-space Episenome unifies cellular information space-time within cognition-based evolution. Prog. Biophys. Mol. Biol. 2020, 150, 112–139.
- Miller, W.B., Jr.; Baluška, F.; Torday, J.S. Cellular senomic measurements in cognition-based evolution. Prog. Biophys. Mol. Biol. 2020, 156, 20–33.
- Tuszynski, J.A.; Portet, S.; Dixon, J.M.; Luxford, C.; Cantielo, H.F. Ionic wave propagation along actin filaments. Biophys. J. 2004, 86, 1890–1903.
- Iglesias, P.A.; Devreotes, P.N. Biased excitable networks: How cells direct motion in response to gradients. Curr. Opin. Cell Biol. 2012, 24, 245–253.
- Pal, D.S.; Li, X.; Banerjee, T.; Niao, Y.; Devreotes, P.N. The excitable signal transduction networks: Movers and shapers of eukaryotic cell migration. Int. J. Dev. Biol. 2019, 63, 407–416.
- Beta, C.; Gov, N.S.; Yochelis, A. Why a large-scale mode can be essential for understanding intracellular actin waves. Cells 2020, 9, 1533.
- Cifra, M.; Pokorný, J.; Havelka, D.; Kucera, O. Electric field generated by axial longitudinal vibration modes of microtubule. Biosystems 2010, 100, 122–131.
- Havelka, D.; Cifra, M.; Kučera, O.; Pokorný, J.; Vrba, J. High-frequency electric field and radiation characteristics of cellular microtubule network. J. Theor. Biol. 2011, 286, 31–40.
- Kučera, O.; Havelka, D. Mechano-electrical vibrations of microtubules—Link to subcellular morphology. Biosystems 2012, 109, 346–355.
- Kalra, A.P.; Eakins, B.B.; Patel, S.D.; Ciniero, G.; Rezania, V.; Shankar, K.; Tuszynski, J.A. All wired up: An exploration of the electrical properties of microtubules and tubulin. ACS Nano 2020.
- Baluška, F.; Reber, A. Senomic and ephaptic principles of cellular consciousness: The biomolecular basis for plant and animal sentience. J. Consc. Stud. 2021, 28, 31–49.
- Tuszynski, J.A. The bioelectric circuitry of the cell. In Brain and Human Body Modeling: Computational Human Modeling at EMBC 2018; Makarov, S., Horner, M., Noetscher, G., Eds.; Springer International Publishing: Basel, Switzerland, 2018; pp. 195–208.
- Tuszynski, J.A.; Friesen, D.; Freedman, H.; Sbitnev, V.I.; Kim, H.; Santelices, I.; Kalra, A.P.; Patel, S.D.; Shankar, K.; Chua, L.O. Microtubules as sub-cellular memristors. Sci. Rep. 2020, 10, 2108.
- Baluška, F.; Volkmann, D.; Barlow, P.W. Nuclear components with microtubule-organizing properties in multicellular eukaryotes: Functional and evolutionary considerations. Int. Rev. Cytol. 1997, 175, 91–135.
