RNAs with methylated cap structures are present throughout multiple domains of life. Given that cap structures play a myriad of important roles beyond translation, such as stability and immune recognition, it is not surprising that viruses have adopted RNA capping processes for their own benefit throughout co-evolution with their hosts. In fact, that RNAs are capped was first discovered in a member of the Spinareovirinae family, Cypovirus, before these findings were translated to other domains of life.
- Spinareovirinae
- reovirus
- capping
- RNA
- Orthoreovirus
- Aquareovirus
- Cypovirus
- transcription
- nucleotide
- virus
1. Introduction
Although this review centers on RNA capping by members of the Spinareovirinae subfamily, we will first provide a brief introduction to the virus family phylogeny, basic structure, and replication cycle.
Phylogeny. Reoviridae is a family of segmented, dsRNA viruses divided into two subfamilies based on the presence of “turrets/spikes” at the vertices of the viral capsid [1]. The non-turreted members of the Reoviridae family form the Sedoreovirinae subfamily, while the turreted viruses form the Spinareovirinae subfamily, the focus of this review. The Spinareovirinae subfamily is composed of nine genera: Orthoreovirus, Aquareovirus, Coltivirus, Mycoreovirus, Cypovirus, Fijivirus, Dinovernavirus, Idnoreovirus, and Oryzavirus, with a wide host range including vertebrates, invertebrates, plants, and fungi. Phylogenetic trees based on the amino acid (aa) sequences of the RNA-dependent RNA polymerases (RdRps) have been built to visualize the relationship between each virus, as depicted in Figure 1. Though structurally conserved, when comparing aa sequences of homologous proteins among the genera, there is often less than 26% sequence similarity. However, protein regions with structural and/or enzymatic function are typically more similar than the rest of the protein [1].
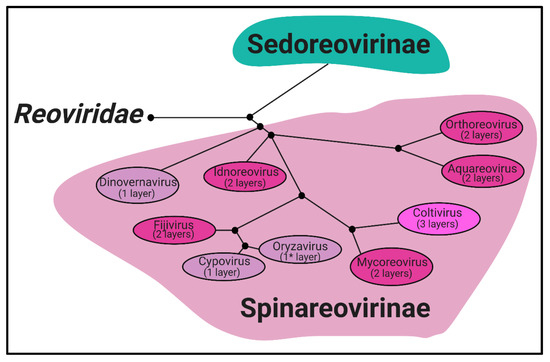
Figure 1.
Spinareovirinae
Reoviridae
Spinareovirinae
Oryzavirus
Mammalian Orthoreovirus
Orthoreovirus
Aquareovirus
Coltivirus
Mycoreovirus
Oryzavirus
Cypovirus
Fijivirus
Idnoreovirus
Dinovernavirus
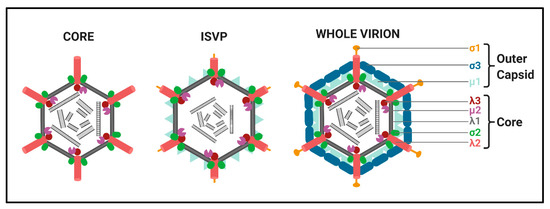
Figure 2.
Mammalian Orthoreovirus (MRV) 3.
Left
Middle
Right
Structure. All Spinareovirinae members share the same, basic virion structure: concentric layers of proteins with icosahedral symmetry encapsidating the segmented dsRNA genomes (Figure 2) [1]. The number of layers varies among the genera, as depicted in Figure 1. All Spinareovirinae members consist of a core, or innermost particle, which is transcriptionally active with T = 1 symmetry. The core particle is formed by 60 asymmetric homodimers of the major-core structural protein, λ1 of Mammalian Orthoreovirus (MRV, or “reovirus” henceforth) [2][3][2,3]. All proteins discussed hereafter will be given the MRV designation; however, it is important to recognize that homologous proteins of the other genera may have different names. Enforcing this backbone structure are the “clamp” σ2 proteins, which are only present among Spinareovirinae [2][4][2,4]. MRV contains 150 copies of the clamp protein, distributed about the 5-, 3-, and 2-fold axes [2]. At the twelve 5-fold axes sits the pentameric “turrets”, formed by the λ2 protein [2][5][2,5]. These turrets form hollow cylinders and possess both methyl- and guanylyl-transferase activity (MTase and GTase, respectively), discussed in detail in later sections. Among all the genera, there are always 12 turrets; however, their shapes and configurations vary. For example, Oryzavirus has both wider and taller turrets compared to MRV [4]. The shape of the λ2 turrets may reflect differences in infection and transcriptional mechanisms; when the outercapsid of MRV is cleaved away, the λ2 turrets take on a more “open” conformation, potentially facilitating the release of mRNA into the host cytoplasm [6].
Some Spinareovirinae members, such as Oryzavirus, Cypovirus, and Dinovernavirus, do not have additional layers beyond the core particle. In contrast, Orthoreovirus, Aquareovirus, Coltivirus, Mycoreovirus, Fijivirus, and Idnoreovirus have layers of outercapsid proteins with considerable structural similarity that surround the innermost core particle, forming a T = 13 symmetrical structure. (Figure 2) [1]. Two major outercapsid proteins, μ1 and σ3, form heterohexamers (200 units for MRV) that attach onto the core particles, forming the T = 13 lattice [3][7][3,7]. The μ1 proteins found within these complexes contact both the σ2 clamps and λ2 turrets to anchor the outercapsid onto the core [2][3][2,3]. In the case of MRV, there is an additional outercapsid receptor binding protein that is absent among the other Spinareovirinae genera, σ1, known to bind both junctional adhesion molecule A (JAM-A) and sialic acids (SAs) on the surface of mammalian cells [1][8][1,8].
Members of the Spinareovirinae subfamily have genomes consisting of 9–12 dsRNA segments, depending on the genera [1]. Most of these dsRNA segments encode a single viral protein, thus they are largely monocistronic. Spinareovirinaes’ genome segments share conserved sequences at their termini with 3′ terminal regions (UCAUC-3′ for MRV) typically more conserved than the 5′ sequences [1]. This may suggest functions for the non-coding regions of reovirus genomes, potentially in the regulation of transcription or encapsidation. As will be discussed in detail throughout this review, the (+)-sense RNAs of Spinareovirinae are capped. Unlike eukaryotic mRNAs, both genomic and transcribed Spinareovirinaes RNAs lack 3′-polyA tails.
Replication Cycle. We will provide a brief overview of the replication cycle for MRV here, as illustrated in Figure 3; however, readers are directed to a recently published review [9] for a more in-depth description. Infection begins with entry of the virus into host cells, a process mediated by the attachment of the σ1 receptor binding protein to sialic acids (SAs) and/or junctional adhesion molecule A (JAM-A) residues [10]. In cell culture, MRVs are generally internalized by clathrin-mediated endocytosis, but can also internalize by phagocytosis or pinocytosis [11]. Within endosomes, acid-dependent proteolytic processing allows stepwise degradation of the outercapsid, beginning with σ3 degradation followed by μ1C cleavage, generating intermediate subviral particles (ISVPs) (Figure 2) [12]. Eventually, transcriptionally active core particles penetrate into the cytoplasm of the cell [12]. Given the natural niche for MRV is the intestine, it is believed that intestinal enzymes such as chymotrypsin kickstart the proteolytic degradation required for infection, generating ISVPs that can penetrate host membranes directly [13]. While MRV entry is well characterized, modes of entry for other Spinareovirinae members likely vary. For example, some Orthoreoviruses express a membrane fusion-inducing protein and can make use of syncytium formation for cell-to-cell spread [14].
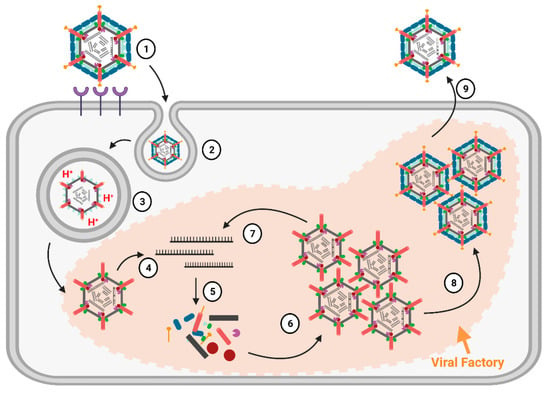
Figure 3.
Mammalian Orthoreovirus 3
An important feature of Spinareovirinae is that RNA transcription occurs within the core particles, and full-length mRNAs are extruded from the λ2 pentameric turrets. As will be comprehensively discussed in this review, the RNAs can also be capped before extrusion to the cytoplasm. The (+)-sense RNAs serve as messenger RNAs for the expression of viral proteins by the host translation machinery. Moreover, as de novo viral capsid proteins are synthesized, (+)-sense RNAs can also be encapsidated into progeny cores. Within progeny cores, (-)-sense RNAs are transcribed, producing the dsRNA genome that serves to amplify (+)-sense RNA synthesis. Primary transcription refers to the first RNA molecules produced by the incoming core particles, while secondary transcription refers to RNA synthesis by newly assembled core particles (Figure 3). The replication of reovirus occurs within localized areas called factories, which are composed of both viral proteins (μ2, μNS, and σNS) and host materials (endoplasmic reticulum (ER) fragments, microtubules, and possibly more) [15]. Ultimately, whole progeny viruses are assembled that can egress from cells, predominantly in a non-lytic manner [9].
2. Capping Status of Viral RNAS
Spinareovirinae +RNAs have Cap1 structures. The years 1974–1976 were riveting for the topic of mRNA capping; Shatkin, Lengyel, and Kozak, along with their trainees and colleagues, discovered that mRNAs were capped and unravelled the enzymatic steps involved in cap addition and modification (Figure 4) [16][17][18][19][20][21][22][23][24][25][26][27][16,17,18,19,20,21,22,23,24,25,26,27]. We recommend visiting these original publications to appreciate how this pivotal discovery was made and disseminated using radioisotopes such as 3H and 32P, enzymatic or chemical treatments, chromatography, brilliant experimental design, and a typewriter. Miura et al. (1974) first discovered that the 5′-terminal phosphates of MRV-synthesized RNAs were in a blocked configuration and could only be labelled by [32P] if the RNAs first underwent oxidation, beta-elimination, and phosphomonesterase treatment [28]. Upon removal of the blocking group, they found that the first nucleotide was always a modified guanine, thought probably to contain a 2′-O-methyl group, followed by a cytosine (GpCp). It is now well recognized that a 5′-GCUA sequence is indeed common to all ten MRV genome segments.
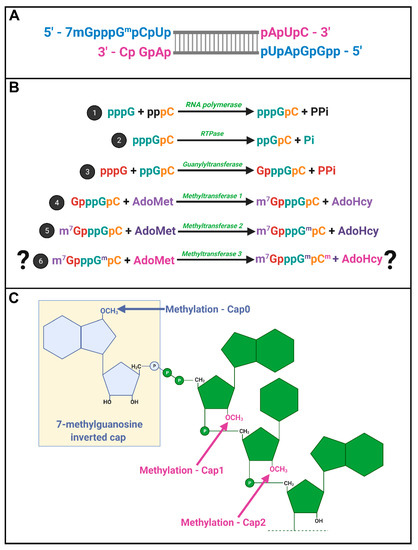
Figure 4.
Spinareovirinae
A
B
Spinareovirinae
C
Motivated by the finding that RNAs of several different viruses, including MRV, also contained methylated nucleotides [29][30][31][32][29,30,31,32], Shatkin’s group set out to define the precise composition of the 5′ termini. Through a series of differential enzymatic treatments and interpretation of chromatographic peaks, the Shatkin group showed that MRV cores synthesize mRNAs with a guanylate cap added to the 5′ end through a 5′-5′ inverted linkage (G(5′)ppp(5′)GpCp), rather than the conventional 5′-3′ linkage. The Shatkin lab also discovered that methyltransferase activities add a methyl group from S-adenosyl-methionine (SAM) to the N7 position of the guanosine cap and the 2′O position of the adjacent first mRNA nucleotide (which is a guanine for MRV) to produce m7G(5′)ppp(5′)Gmp; a structure later termed “cap1”. It was already known that a viral-associated RNA triphosphatase (RTPase) could remove the γ-phosphate of nucleotides [33][34][33,34]; this hydrolase activity was now predicted to produce the diphosphate 5′guanine-cytosine terminus (5′ppGpCp) to which the inverted guanosine cap is added. One month following their description of the cap1 structure on in vitro synthesized RNAs, the Shatkin team demonstrated that the same cap1 structure was found on RNA genomes of viruses purified from infected cells; this suggested that cap1 structures are indeed produced during MRV infection and not just an artifact of in vitro systems [25]. By 1976, the cap1 structure was observed in numerous viruses and eukaryotes ranging from humans to silkworms to yeast; mRNAs being capped became an accepted dogma. The Spinareovirinae genomic dsRNA was then found to consist of capped (+)-sense RNAs, but diphosphate bearing (−)-sense RNAs that are uncapped and unmethylated at the 5′ end (Figure 4A).
In 1976, Furuichi et al. depicted the sequence of capping reactions along with the substrates and enzymatic activities involved [16][17][20][16,17,20]. Specifically, RTPase, guanylyltransferase (GTase), and two methyltransferases (MTases) are necessary, in that order, to generate the final cap1 structure on reovirus RNAs (Figure 4B). Later sections will discuss the possible location and orchestration of these enzymes in the context of Spinareovirinae.
Does reovirus also make cap2 structures? In 1976, Desrosiers et al. found that in L929 cells, 50% of MRV derived mRNAs also had a methyl group on the 2nd nucleotide not counting the guanosine cap; a structure referred to as cap2 (m7G5′ppp5′GmpCmp) (Figure 4C) [35]. The presence of cap2 structures on reovirus RNAs was also suggested by Shatkin and Both [19]. Specifically, while (+)-sense RNAs generated in vitro by reovirus cores had cap1, ~40% of (+)-sense RNAs synthesized during the infection of L929 cells at 5–11 h post infection (hpi) had cap2 structures. Moreover, cap2 was absent from the (+) strand of the dsRNA genome, suggesting that packaged (+)RNA was cap1-modified while ~40% of unpackaged (+)RNAs had cap2 structures [36]. Based on these findings, the authors proposed that cap1 versus cap2 structures may help regulate the fate of (+)RNAs, distinguishing protein translation versus packaging.
3. Functions of RNA Capping during Infection
Viral RNA transcription is not contingent on capping activity. As early as 1975 when the 5′cap structure was being discovered, there were speculations about its possible functions. Early studies demonstrated that capping was not essential for viral RNA transcription. Specifically, Reeve et al. [37][44] showed that the addition of [γ-S]GTP effectively prevented capping by resisting hydrolysis to the diphosphate (ppGpCp) substrate required for the capping reaction. Importantly, the addition of [γ-S]GTP did not interfere with viral RNA synthesis in vitro [37][44]. It would be interesting to perform similar studies in cell culture infections, not only to confirm that capping is dispensable for viral RNA synthesis, but as a strategy to explore the role of capping during reovirus replication. Noteworthy is that although [γ-S]GTP did not prevent RNA transcription, this does not indicate whether the RNA template within the virus particle had to be capped to serve as a template, as the addition of [γ-S]GTP does not affect the capping status of the input genomic template.
RNA capping promotes virus protein translation and RNA assembly. Unlike for viral RNA synthesis, the importance of mRNA capping for protein translation was well established by the 1980s. MRV mRNAs produced in the presence of methyl-donor SAM exhibited heightened ribosome binding and efficient translation in wheat germ or L929 cell lysate-based cell free translation systems [21][38][21,45]. These early studies inferred that 5′caps are involved in ribosome recruitment. Marilyn Kozak and Aaron Shatkin then demonstrated in 1976 that, indeed, the 43S ribosome initiation complex was recruited to the 5′terminus of mRNAs [18]. A higher proportion of 43S-protected fragments was obtained using methylated versus unmethylated mRNA, suggesting that m7G contributes to increased efficiency of 43S binding but is not absolutely required [38][45]. Furthermore, the 80S ribosomal complex was suggested to be formed at AUG-containing regions by recruitment of the 60S ribosomal subunit, since cap-containing RNAs without AUG start codons showed neither 43S nor 80S ribosome rebinding.
Given that mRNA capping promotes translation by recruiting the 43S ribosome complex to the initiation AUG site, the next logical question to ask would be: to what extent does the capping of reovirus mRNA enhance protein translation and assist in competition for host translation machinery? To begin addressing this question, our lab recently used the reovirus reverse genetics approach to compare de novo virus protein synthesis and progeny production in the presence versus absence of mRNA capping [39][46]. Specifically, transcription of all ten plasmid-derived MRV genome segments was driven by a T7 RNA polymerase promoter as is typical for the reverse genetics system, but the baby hamster kidney (BHK) cells were also variably transfected with the African Swine Fever Virus NP868R capping enzyme. In these experiments, it is important to point out that host capping enzymes are normally found in the nucleus where they remain inaccessible to reovirus particles present in the cytoplasm, thus the transfection of an exogenous capping enzyme allows mRNA produced in the cytoplasm to be capped. With that in mind, the inclusion of NP868R increased reovirus protein levels by ~10-fold, yet increased new progeny particle production by ~100-fold. These experiments suggested that mRNA capping serves additional functions beyond protein expression. To determine if capping promotes translation-independent steps of reovirus replication, BHK cells were infected with reovirus to produce capped viral RNAs, but also transfected concurrently with a plasmid-derived S1 genome segment modified to encode the green fluorescent protein, UnaG. Importantly, the experiments were conducted in the presence or absence of NP868R to produce capped or uncapped S1-UnaG mRNAs, respectively. In this experimental system, the virus infection produced all components for virus amplification and assembly, and the fate of capped or uncapped S1-UnaG alongside the infection could be monitored. Purified virions from the infected-transfected BHK cells were then added to Ras-transformed NIH3T3 cells, and the expression of UnaG from progeny virions was assessed by RT-qPCR and flow cytometry. S1-UnaG expression was only observed when progeny virions were produced in the presence of NP868R. These findings can be interpreted in two ways: (1) capping was essential for S1-UnaG mRNA to be encapsidated, or (2) both capped and uncapped S1-UnaG mRNAs were encapsidated but capping was essential for the subsequent transcription and expression of S1-UnaG transcripts. As discussed below, the reovirus polymerase has a cap-binding site that was previously suggested to serve as an anchor for the viral genomic RNA during transcription. Thus, a feasible possibility to explain the lack of infectious progeny with uncapped S1-UnaG is that the association of the polymerase with RNA caps is essential for RNA encapsidation, transcription, or both.
Aside from its roles in virus replication, the composition of the 5′ termini of RNAs can affect the host response to virus infection. It is now well established that viral RNAs are recognized as foreign by pattern recognition receptors such as toll-like receptors (TLR), RIG-I–like receptors (RIG-I), double-stranded RNA-activated protein kinase (PKR), and melanoma differentiation–associated protein 5 (MDA5). The binding of viral RNA to these receptors results in type I IFN induction and downstream expression of IFN-stimulated genes (ISG), many of which have antiviral properties. RIG-I was found to be the key pattern recognition receptor during MRV infection [40][47], as shRNA-mediated silencing of RIG-I prevented IFN production following reovirus infection. Additionally, while Ras-transformed cells with impaired RIG-I signaling permitted efficient reovirus dissemination, the silencing of RIG-I in non-transformed cells with functional RIG-I-signaling was necessary to allow efficient MRV cell-to-cell spread. In these studies, either MRV infection or transfection of in vitro synthesized m7G-capped but unmethylated reovirus (+)RNAs induced IFN-dependent antiviral effects. It would be interesting to repeat these studies with differentially modified reovirus (+)RNAs, cap1 or cap2 structures, to determine the role of methylation for RIG-I detection. Another study showed that 5′diphosphate-bearing reovirus -RNAs were also able to induce the IFN response in a RIG-I-dependent manner [41][48].
