Saccharomyces cerevisiae has been for a long time a common model for fundamental biological studies and a popular biotechnological engineering platform to produce chemicals, fuels, and pharmaceuticals due to its peculiar characteristics. Both lines of research require an effective editing of the native genetic elements or the inclusion of heterologous pathways into the yeast genome. Although S. cerevisiae is a well-known host with several molecular biology tools available, a more precise tool is still needed. The clustered, regularly interspaced, short palindromic repeats–associated Cas9 (CRISPR-Cas9) system is a current, widespread genome editing tool. The implementation of a reprogrammable, precise, and specific method, such as CRISPR-Cas9, to edit the S. cerevisiae genome has revolutionized laboratory practices.
- Saccharomyces cerevisiae
- CRISPR-Cas9
- CRISPRi
- CRISPRa
- dCas9
- CRISPR-ARE
- CRISPR-AID
1. Introduction
Saccharomyces cerevisiae has long been a common model for fundamental biological studies because of its safety and ease of handling. This yeast has been extensively used to elucidate eukaryotic processes from basic metabolism to protein/gene interaction or even evolution [1]. Moreover, it is also a popular biotechnological engineering platform used to produce chemicals, fuels, and pharmaceuticals [2]. Associated with these two lines of research is the need to effectively edit the native genetic elements or introduce heterologous pathways into the yeast genome. For instance, for metabolic engineering purposes, several genetic modifications are required besides the introduction of heterologous genes in order to stream the metabolic fluxes toward the production of the desired product. The development of an efficient cell factory is associated with several native gene deletions, overexpression, or replacements. All these modifications require a cycle of individual transformation, selection, and confirmation, thus making the process very time-consuming. Furthermore, each cycle is associated with the integration of a selective marker. However, the number of markers available is limited, which also limits the number of sequential modifications that can be performed.
S. cerevisiae is known to possess a very efficient homologous recombination (HR) machinery [3]. Researchers have been taking advantage of this feature for in vivo assembly of multiple linear fragments instead of using in vitro molecular biology techniques. Most of the in vivo assembly performed in yeast was focused on circular vector construction. However, for metabolic engineering purposes, genomic integration offers a more stable expression system and eliminates the need of ensuring a selective pressure for plasmid maintenance. Nevertheless, in vivo assembly in combination with genomic integration is associated with very low efficiencies [4].
Double-strand breaks (DSBs) in the genome occur due to several environmental factors and can be repaired by HR or nonhomologous end-joining (NHEJ). In S. cerevisiae it is known that the dominant repair is performed by HR. The introduction of a DSB has been shown to increase the efficiency of homologous integration of linear DNA fragments with homologous ends [5]. These features led researchers to develop methods that use DSBs for site-directed genome editing. Theoretically, these methods could be used for marker-free modifications and examples include zinc finger nucleases (ZFN) and transcription-activator-like effector nucleases (TALEN). Both methods involve the design of proteins that recognize a specific DNA sequence and cause a site-specific DSB through DNA–protein interaction. The DSB is then repaired by HR with a provided DNA fragment with homologous ends. However, a new ZFN or TALEN protein has to be engineered for each target modification, making these methods also very laborious [6][7].
Evolution allowed bacteria to develop several systems to degrade foreign DNA as a defense mechanism. The most well-known are restriction enzymes, which rapidly became the “workhorses of molecular biology” [8]. More recently, another prokaryotic immune system was identified [9]. This system consists of clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated nucleases (CRISPR-Cas), widely distributed among bacteria and archaea [10]. CRISPR-Cas has the function of recognizing and degrading invading nucleic acids. The system is divided in three classes (I, II, and III) based on the cas gene sequences, operon organization, and the repeats within CRISPR arrays [11]. The increased knowledge of CRISPR action mechanism broke new ground regarding a possible applicability for this system to edit genetic elements of an organism.
Due to their simplicity relative to other classes, class II CRISPR-Cas systems have the most well-developed methods for genomic engineering nowadays. In class II, all the functions of the effector complex are performed by a single protein [12]. The following components are required: the RNA-guided nuclease Cas9, a CRISPR RNA (crRNA), an auxiliary trans-activating crRNA (tracrRNA), and an RNase III. The action mechanism is simple and aims at the creation of a DSB in the target DNA. A hybrid of the RNA molecules directs Cas9 to the target DNA site and the Cas9 cleaves a targeting DNA sequence containing a protospacer-adjacent motif (PAM) sequence. Jinek et al. [13] postulated that the requirement for RNase and tracRNA can be bypassed by fusion crRNA and tracrRNA to form a single-guide RNA (gRNA) simplifying the process and application. The gRNAs are composed of a homologous sequence 20 nt upstream of the PAM sequence (guide sequence) and the scaffolding loop structure to attach the Cas9 (Figure 1).
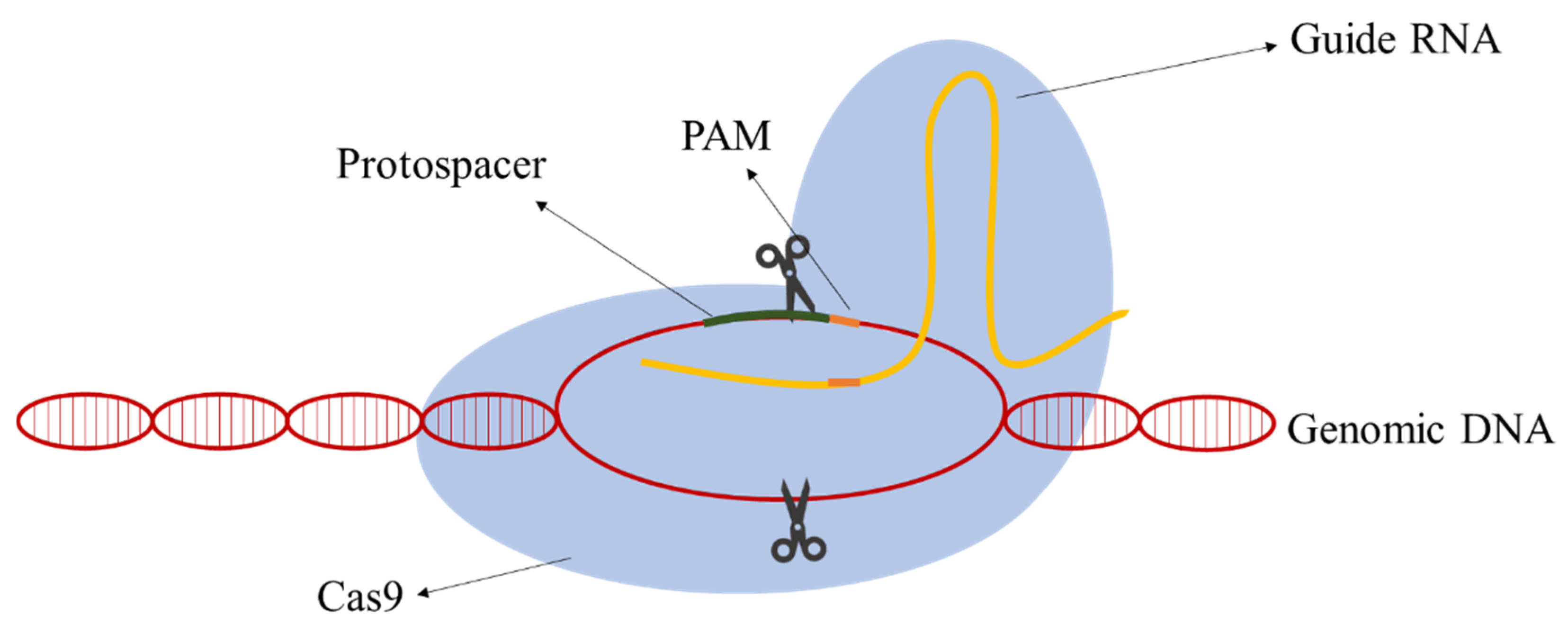
The type II CRISPR-Cas9 system was implemented in S. cerevisiae for genetic modification purposes by DiCarlo et al. [14]. The researchers demonstrated that co-transforming a gRNA targeting a negative selectable marker together with a 90-bp double-stranded HR donor with a frameshift mutation in the targeted reading frame and PAM replacement by a stop codon resulted in almost 100% of mutated cells.
2. Innovative CRISPR Toolkits in Saccharomyces cerevisiae
New emergent procedures into how CRISPR-Cas9 is applied have recently been developed demonstrating the potentialities of this tool. The target specificity of CRISPR-Cas9 systems allowed researchers to develop technologies that take advantage of its precise DNA targeting namely for the delivery of other molecules. EvolvR, for instance, combines the target specificity of CRISPR-Cas9 technology with the error-prone capacity of a mutant DNA polymerase for in vivo targeted nucleotide diversification [15]. The system employs the use of a nicking variant of Cas9 (nCas9) that cuts only one DNA strand avoiding native homology repair, and the DNA polymerase uses the nick as a start point to initiate mutation insertion. This technology was recently applied in S. cerevisiae, yEvolvR [16]. The results demonstrated that yEvolvR was able to insert random mutations in both directions of the target sequence, and in addition, it was possible to target two genomic loci at the same time. This technology could be important for fundamental eukaryotic research such as protein function or protein interactions or to investigate genetic mechanisms, where yeast is usually used as a role model. Moreover, it could be applied to strain tolerance engineering for industrial purposes. Engineering yeasts to confer them with increased tolerance to an external stress, such as temperature or oxidation, is a very valuable strategy to improve industrial strains’ performance. CRISPR has already been implemented for random mutagenesis via genome shuffling. Mitsui et al. [17] applied CRISPR-Cas9 for cleaving the δ-sequences in order to fragment the chromosome. During the repair of DNA fragments, large-scale modifications, such as gene amplification, translocation, and deletion, may occur. In this case, the DNA repair was induced under thermal stress conditions. After DNA repair, the modified yeast was able to grow at 39 °C and reported higher ethanol and acid resistance than the parental strain.
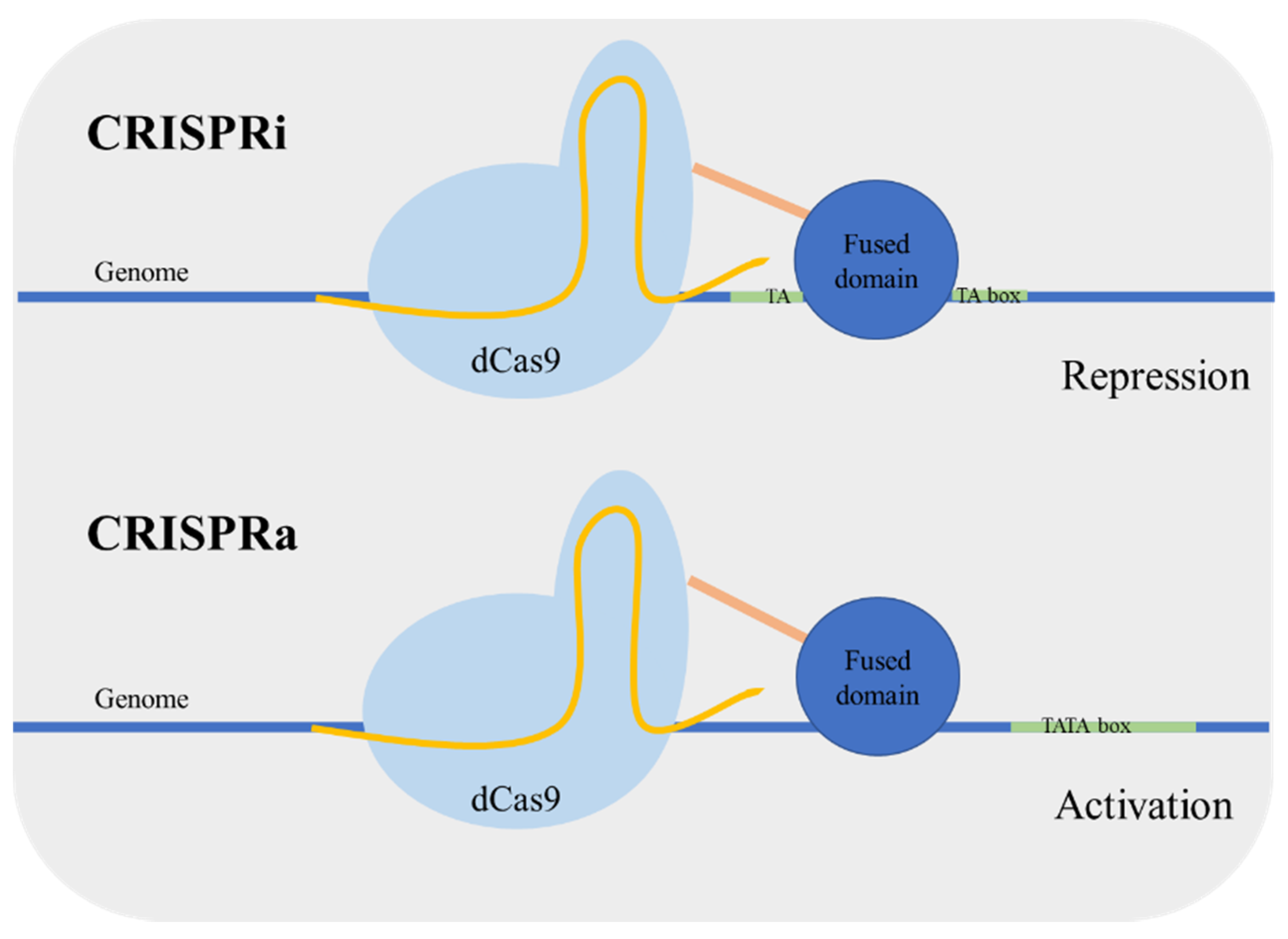
Targeted regulation of gene expression is important both in the context of metabolic engineering and functional genomics [18]. In yeast, the control of genetic expression is usually performed using characterized gene promoters with different strengths; however the prediction of the expression level remains a challenge. Qi et al. [19] developed an enzymatic version of Cas9 mutated in the nuclease nucleotide sites designated as dead Cas9 (dCas9). This Cas9 mutant is defective in DNA cleavage, and it can act as a simple specific DNA binding complex. Using this version of Cas9 for targeting a coding sequence caused transcriptional gene repression in Escherichia coli. The dCas9 binds to the target sequence blocking the action of RNA polymerase. This approach was named CRISPR interference (CRISPRi). Next, the same research group applied the system for gene repression in S. cerevisiae [20]. The dCas9 was guided to a specific promoter, resulting in an efficient gene repression. In addition, the repression can be enhanced by fusing a transcriptional repressor domain to dCas9. Alternatively, Farzadfard et al. [21] fused dCas9 to an activator domain and reported an activation or repression depending on the targeting site. When the target was outside the TATA box, the promoter was activated (CRISPR activation (CRISPRa)), targeting adjacent to the TATA box resulted in gene repression (CRISPRi) (Figure 2). Other activator-domain-fused dCas9 proteins were also developed achieving higher regulation levels [22]. Moreover, the CRISPRa/i system has been applied in a polyploid yeast strain, which can be very valuable for the development of more robust industrial stains [23].
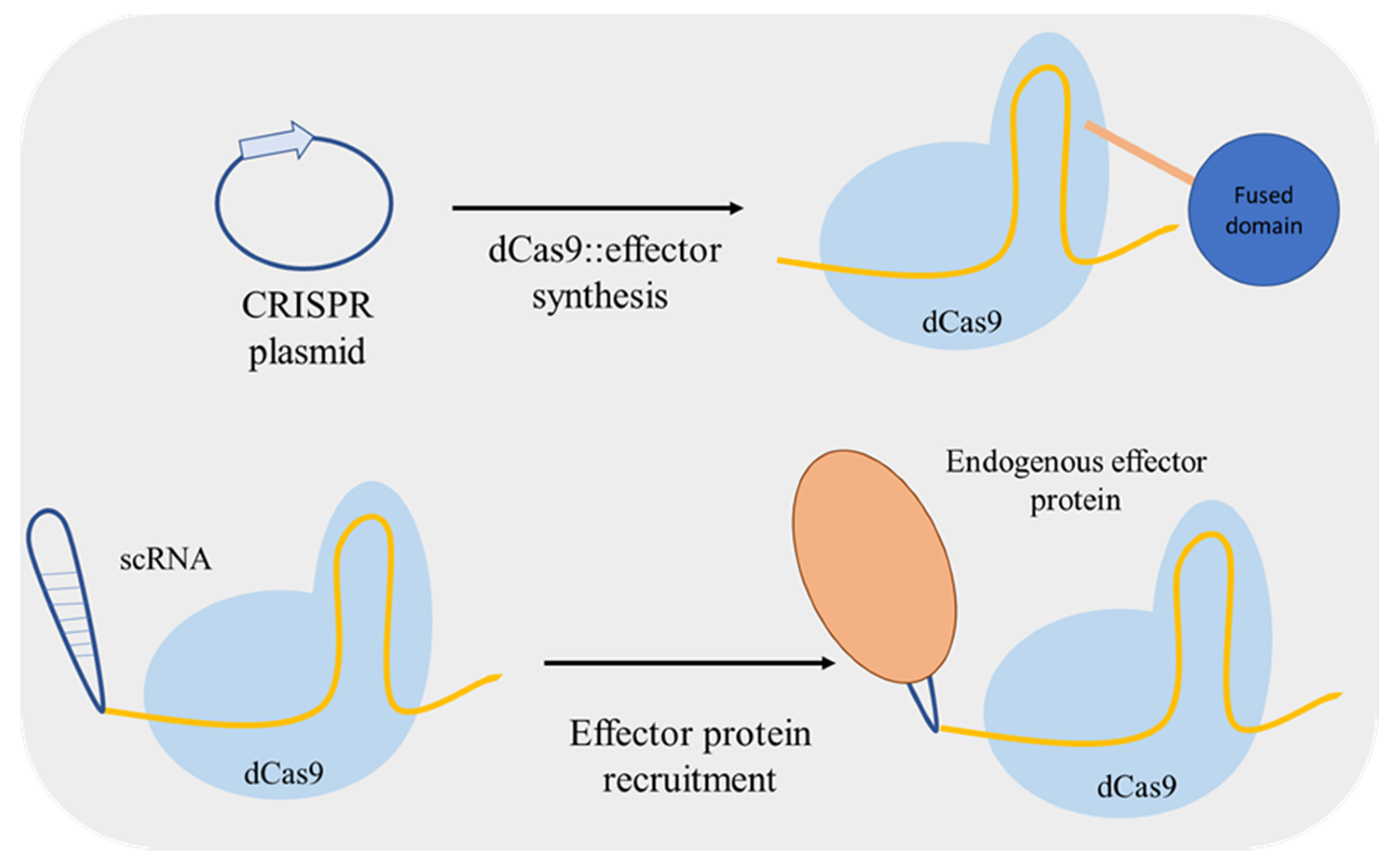
Zalatan et al. [24] used a different approach for up/downregulation of a target gene. Instead of including fusion domains in dCas9, the authors included effector protein recruitment RNA domains into the gRNA converting it to a scaffold RNA (scRNA) (Figure 3). The RNA hairpins of scRNA can recruit a specific RNA-binding protein, an activator or a repressor, thus used for locus-specific regulation. Furthermore, Jensen et al. [25] compared the regulatory performance of two distinct dCas9-mediated systems: using an inducible gRNA expression and dCas9 fused with a repressor or an activator domain; and with constitutive expression of scRNAs for effector molecule recruiting. The two systems mediated similar changes in activation/repression of the targeted promoters both at single and at multiplex level.
Moreover, a grade modulation of genetic expression was reported by Deaner and Alper [26]. The range of genetic expression was related to the proximity of dCas9-based regulators to the core of the promoter. The grade modulation applicability represents a step forward to enable a fine tuning of metabolic pathways in S. cerevisiae. The possible applications of CRISPRi/a are massive, and they have already been used to improve a yeast cell factory to produce β-amyrin [27].
Another possible approach, besides the use of a modified CRISPR protein, to guide effector molecules without creating a DSB, is by adjusting the gRNA molecule length. Truncated gRNAs, usually of 14 nt, proved to be able to guide the binding of Cas9 to the target sequence without the introduction of a DSB [28][29]. Hereupon, truncated gRNAs can be used for transcriptional regulation and, simultaneously, full-length gRNAs can be used for genome editing using a single Cas9 protein. This feature allowed researchers to develop multifunctional systems capable of one-pot CRISPRi, CRISPRa, or CRISPR editing. Lian et al. [30] reported the first trifunctional CRISPR system for simultaneous gene inactivation, activation, and editing in S. cerevisiae using truncated gRNAs for CRISPRa and CRISPRi, called CRISPR-AID. However, to avoid competition between gRNAs for the same Cas9 protein, three different PAM-recognizing CRISPR proteins were used. More recently, Dong et al. [31] established a trifunctional CRISPR system using a single Cas9 protein. The system was named CRISPR-ARE and employed the use of a Cas9 fused to a VP64-p65-Rta (VPR) activation domain. The authors demonstrated the applicability of CRISPR-ARE by optimizing α-santalene biosynthesis. The system used truncated gRNAs to target the genes for activation and repression and full-length gRNAs to edit one gene by transforming together the donor repair DNA. The editing efficiency was 100% and, gene activation and gene repression were confirmed using a reporter protein. Regarding the α-santalene biosynthesis, it increased 2.66-fold.
References
- Botstein, D.; Fink, G.R. Yeast: An experimental organism for 21st century biology. Genetics 2011, 189, 695–704.
- Nielsen, J.; Larsson, C.; van Maris, A.; Pronk, J. Metabolic engineering of yeast for production of fuels and chemicals. Curr. Opin. Biotechnol. 2013, 24, 398–404.
- Bernardi, B.; Wendland, J. Homologous Recombination: A GRAS Yeast Genome Editing Tool. Fermentation 2020, 6, 57.
- Kuijpers, N.G.A.; Solis-Escalante, D.; Bosman, L.; van den Broek, M.; Pronk, J.T.; Daran, J.M.; Daran-Lapujade, P. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb. Cell Fact. 2013, 12, 1–13.
- Kuijpers, N.G.A.; Chroumpi, S.; Vos, T.; Solis-Escalante, D.; Bosman, L.; Pronk, J.T.; Daran, J.M.; Daran-Lapujade, P. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 769–781.
- Rodrigues, J.L.; Rodrigues, L.R. Synthetic Biology: Perspectives in Industrial Biotechnology. In Foundations of Biotechnology and Bioengineering, Volume 1—Current Developments in Biotechnology & Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 239–269.
- Rodrigues, J.L.; Ferreira, D.; Rodrigues, L.R. Synthetic biology strategies towards the development of new bioinspired technologies for medical applications. In Bioinspired Materials for Medical Applications; Elsevier: Amsterdam, The Netherlands, 2017; pp. 451–497.
- Roberts, R.J. How restriction enzymes became the workhorses of molecular biology. Proc. Natl. Acad. Sci. USA 2005, 102, 5905–5908.
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712.
- Luo, M.L.; Leenay, R.T.; Beisel, C.L. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 2016, 113, 930–943.
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477.
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- DiCarlo, J.; Norville, J.; Mali, P.; Rios, X.; Aach, J.; Church, G. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343.
- Halperin, S.O.; Tou, C.J.; Wong, E.B.; Modavi, C.; Schaffer, D.V.; Dueber, J.E. diversification of all nucleotides in a tunable window. Nature 2018, 560, 248–252.
- Tou, C.J.; Schaffer, D.V.; Dueber, J.E. Targeted diversification in the Saccharomyces cerevisiae genome with CRISPR-guided DNA polymerase I. ACS Synth. Biol. 2020, 9, 1911–1916.
- Mitsui, R.; Yamada, R.; Ogino, H. Improved Stress Tolerance of Saccharomyces cerevisiae by CRISPR-Cas-Mediated Genome Evolution. Appl. Biochem. Biotechnol. 2019, 189, 810–821.
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Res. 2017, 17.
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183.
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. XCRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442.
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604–613.
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328.
- Cámara, E.; Lenitz, I.; Nygård, Y. OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6–12. Sci. Rep. 2020, 10, 1–13.
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015, 160, 339–350.
- Jensen, E.D.; Ferreira, R.; Jakočiūnas, T.; Arsovska, D.; Zhang, J.; Ding, L.; Smith, J.D.; David, F.; Nielsen, J.; Jensen, M.K.; et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb. Cell Fact. 2017, 16, 1–16.
- Hassing, E.J.; de Groot, P.A.; Marquenie, V.R.; Pronk, J.T.; Daran, J.M.G. Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae. Metab. Eng. 2019, 56, 165–180.
- Yu, Y.; Chang, P.; Yu, H.; Ren, H.; Hong, D.; Li, Z.; Wang, Y.; Song, H.; Huo, Y.; Li, C. Productive Amyrin Synthases for Efficient α-Amyrin Synthesis in Engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2018, 7, 2391–2402.
- Kiani, S.; Chavez, A.; Tuttle, M.; Hall, R.N.; Chari, R.; Ter-ovanesyan, D.; Qian, J.; Pruitt, B.W.; Beal, J.; Vora, S.; et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 2015, 12, 1051–1054.
- Dahlman, J.E.; Abudayyeh, O.O.; Joung, J.; Gootenberg, J.S.; Zhang, F.; Konermann, S. Orthogonal gene knockout and active Cas9 nuclease. Nat. Biotechnol. 2015, 33, 3–5.
- Lian, J.; Hamedirad, M.; Hu, S.; Zhao, H. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 2017, 8, 1–9.
- Deaner, M.; Alper, H.S. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab. Eng. 2017, 40, 14–22.
